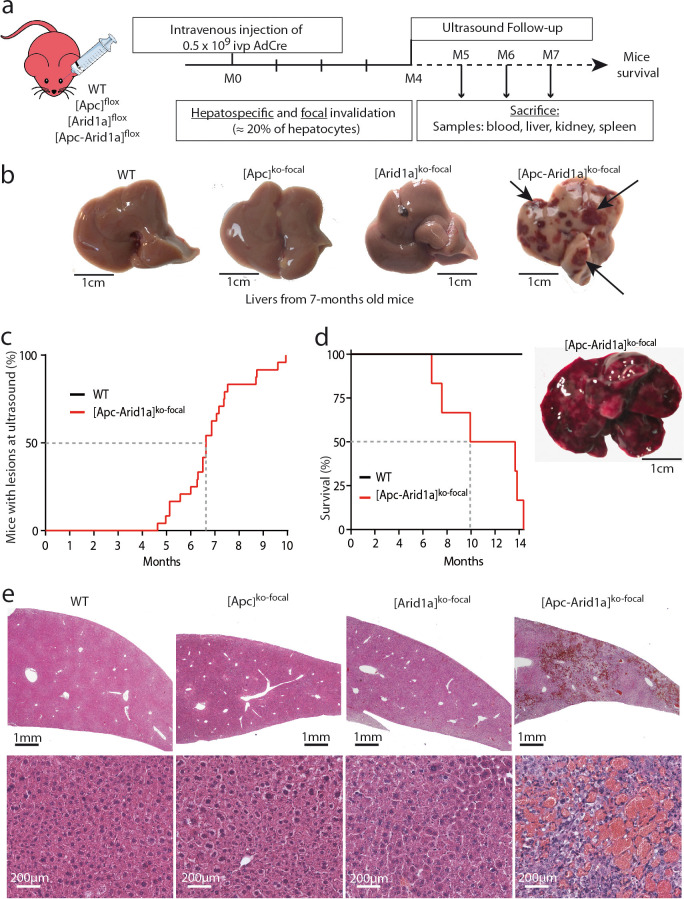
Figure 1. Development of peliosis-like regions after hepato-specific and focal Arid1a and Apc inactivation.
(a) Cre-loxP-generated hepatocyte-specific and inducible inactivation of Apc and/or Arid1a in 20% of hepatocytes after retro-orbital injection of infectious viral particles (ivp) of adenovirus encoding Cre recombinase (AdCre). The resulting mice are referred to as [Apc-Arid1a]ko-focal, [Apc]ko-focal, and [Arid1a]ko-focal. (b) Gross examination of mouse livers, 7 months after AdCre injection. Livers from [Apc-Arid1a]ko-focal mice had an irregular shape and a rough surface, with multiple dark red zones (indicated by arrows). (c) Incidence of hepatic lesions detected in WT (n = 10) and [Apc-Arid1a]ko-focal (n = 24) mice by ultrasonography. (d) Kaplan-Meier estimated survival curves of WT and [Apc-Arid1a]ko-focal mice over 15 months. n = 6 for each group. Inset: Liver of one mouse at necropsy (13 months after AdCre injection, representative of the three analyzed mice). (e) Hematoxylin Eosin (HE)-stained sections of mouse livers at 7 months post-injection. Large vascular spaces filled with blood cells were observed only in [Apc-Arid1a]ko-focal livers. Related data are found in Figure 1—figure supplements 1–4, and source data in ‘Figure 1—source data 1; Figure 1—figure supplement 1—source data 1; Figure 1—figure supplement 3—source data 1’.