Figure 3.
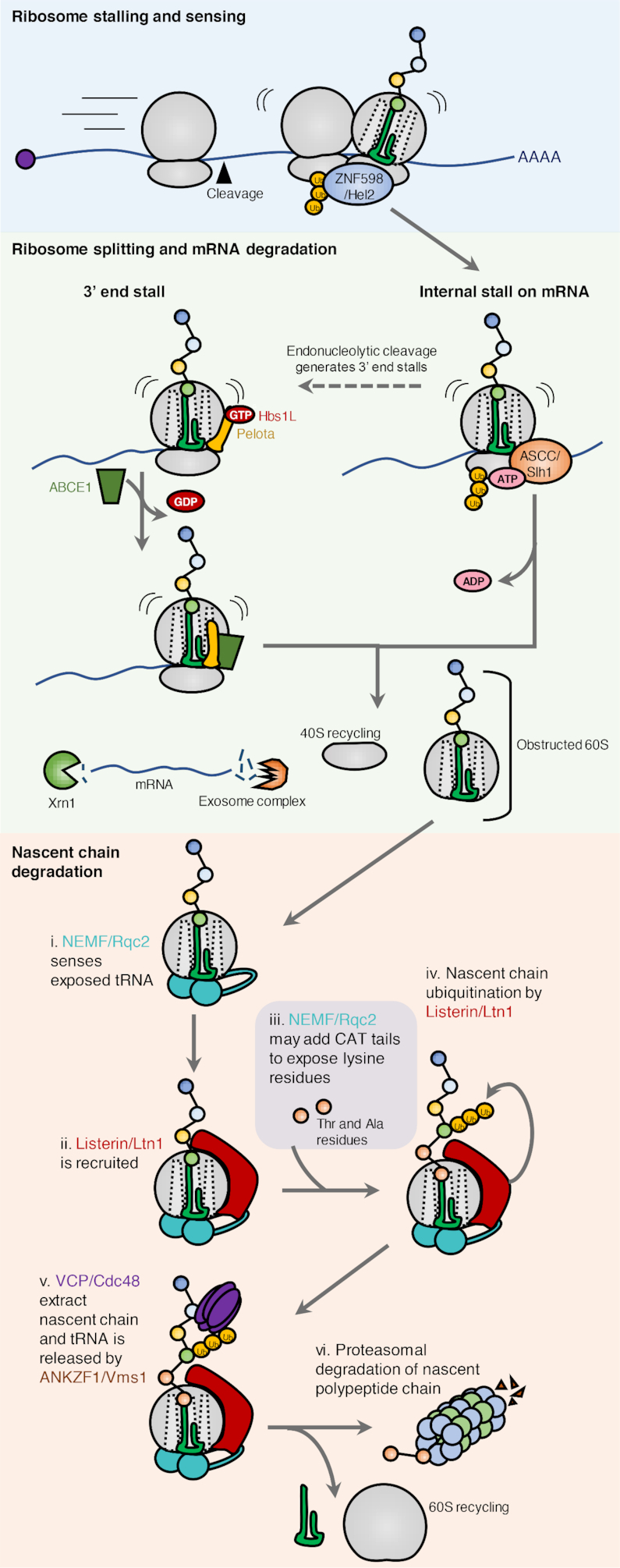
Ribosome collision and ribosome-associated quality control. (A) Translational stalling causes collision of ribosomes. This unique structure is recognized by the E3 ubiquitin ligase ZNF598 that promotes ubiquitination of the ribosomal proteins uS10, eS10 and uS3. Stalling on internal mRNA sequences is recognized by the ASC complex (Slh1 in yeast) which liberates the leading ribosome. The trailing ribosomes can then resume translation. Under certain circumstances, endonucleases cleave the mRNA between ribosomes resulting in ribosomes stalled on 3′end of the mRNA. This makes them accessible for splitting by the recycling factors Pelota, HBS1L and ABCE1 (Dom34, Hbs1 and Rli1 in yeast). The released mRNA is degraded by the 5′-3′ exoribonuclease Xrn1 and the exosome complex to prevent the aberrant mRNA from being translated again. While the 40S subunit is directly ready for recycling, the peptidyl-tRNA remains associated with the large ribosomal subunit. The obstructed 60S subunit is recognized by the RQC component NEMF (Rqc2 in yeast) that recruits the E3 ubiquitin ligase Listerin (Ltn1 in yeast) to the native peptide chain. NEMF/Rqc2 may employ ‘CAT-tailing’ to expose ribosome-buried and ubiquitinatable lysine residues in the native chain. Ubiquitination by listerin/Ltn1 recruits the ATPase VCP (Cdc48 in yeast), and once the nascent chain has been released from the tRNA by ANKZF1 (Vms1 in yeast), VCP can deliver the polypeptide to the proteasome, where it is degraded.
