Figure 5.
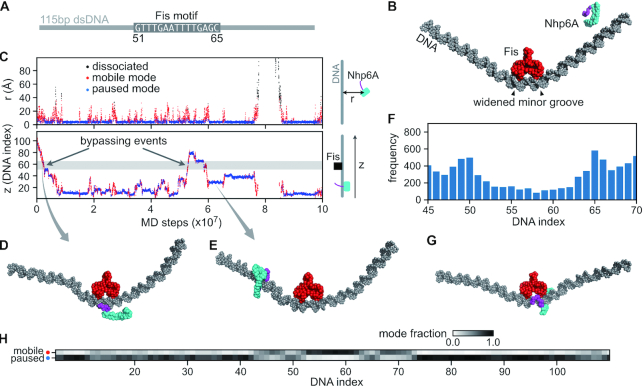
Molecular dynamics (MD) simulations of Nhp6A bypassing a stationary Fis dimer. (A) A 115 bp dsDNA was used in the Nhp6A+Fis+DNA coarse-grained MD simulations. The central 15 bp was the Fis consensus motif (from DNA index 51–65), and the other regions had random sequences. (B) Initial structure of the MD simulations. Nhp6A was placed near one end of the DNA. The N-terminal arm of Nhp6A is shown in purple and the folded HMGB domain is in cyan. (C) A representative MD trajectory of Nhp6A sliding on DNA. Top panel: time series of distance from center-of-mass of Nhp6A HMGB domain to DNA (r); Bottom panel: Nhp6A binding position on DNA (z). In both panels the dots are colored by their corresponding binding status: mobile (red) or paused (blue) binding modes or dissociated from DNA (black). The grey region in the bottom panel represents the Fis binding motif (51–65). (D) A representative structure of Nhp6A using its N-terminal arm to contact DNA during Fis bypass. (E) A representative structure of Nhp6A binding to a Fis-free region in the paused mode. f) Nhp6A binding frequency to the region 45–70 on DNA in 50 independent MD simulations. (G) A representative structure of Nhp6A binding to the widened minor groove at DNA index 65. h) Normalized fraction of the two binding modes of Nhp6A along the DNA.