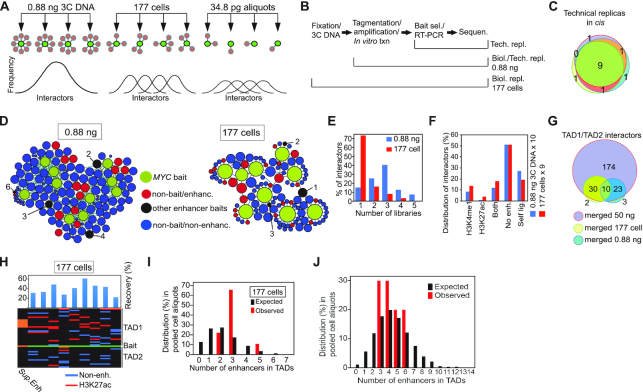
Figure 2.
The strategy to discriminate between virtual and real enhancer hubs. (A) The principle of analysis of stochastic chromatin networks displayed in libraries with smaller amount of input. The frequency of the distribution of interactors in the panel showing the 0.88 ng aliquots are predicted to closely follow a normalized distribution profile representative of the initial 3C sample derived from one million cells. Under the assumption that the network represents stochastic interactions, the biological variability is expected to be higher in the 177-cell sample than in the 0.88 ng sample with the highest variability represented by the 34.8 pg/21 alleles samples. (B) Sampling of technical and biological replicas. ‘Tech.’ represents technical whereas ‘Biol.’ represents biological replicas. (C) Venn diagram showing the overlap in interactors in cis between three different technical replicas. (D) Chromatin networks detected by the MYC bait in 10 samples of 0.88 ng 3C DNA aliquots (average recovery was 74.4%) or 9 different samples each containing 177 cells. The size of each node reflects its connectivity. (E) Bar diagram showing the relationship between the number of interactors (in cis) versus the number of the replicates showing the specific interaction. The bait was omitted from this analysis. (F) The stratification of interactors based on the presence or absence of enhancer marks in HCT116 cells. (G) Venn diagram demonstrating the overlap between pooled small input samples and the 50 ng ‘ensemble’ library. (H) Heatmap of enhancer-MYC interactions in the flanking TADs within the libraries of the 177 cell aliquots. Black indicates absence of interacting sequences. (I) The observed frequencies of enhancers impinging on the MYC bait compared to the expected frequencies, generated by random resampling of interactors in TADs with enhancer marks impinging on the MYC bait scaled from 50 ng 3C DNA input to 177 cells. (J) The observed frequencies of enhancers impinging on the MYC bait compared to the expected frequencies, generated by random resampling of interactors in TADs scaled from 50 ng 3C DNA input to 0.88 ng. Values were calculated from interactors in the whole genome. A statistical test showed that the P-values for panels I and J were found to be 0.3274 and 0.6373, respectively. In both instances it thus failed to reject the null hypothesis at the 95% confidence level.