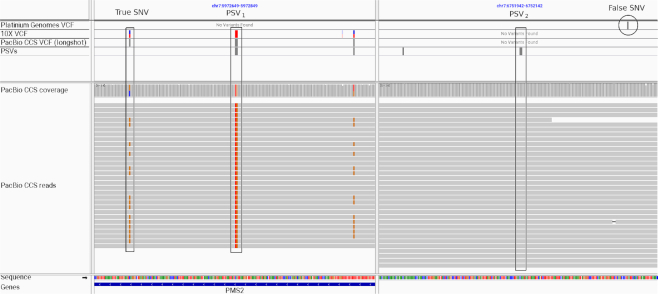
Figure 6.
Illustration of how unreliable PSVs adversely impact short-read variant calling in segmental duplications. An Integrated Genomics Viewer view of a duplicated region on chromosome 7 that overlaps the PMS2 gene is shown. A PSV is located at chr7:5972749 (allele = with the homologous position at chr7:6752042). Variant calling on PacBio CCS reads (aligned with DuploMap) identifies two variants, a homozygous variant at the PSV site (chr7:5972749:CA:TG) and a heterozygous SNV located nearby (chr7:5972674:C:G). Both of these variants are supported by 10X Genomics variant calls but are absent from short-read variant calls for the same individual (PG VCF track). In addition, short-read variant calling results in a false SNV (chr7:6752118:G:C) at the position homologous to chr7:5972674—a result of short-read mismapping due to the unreliable PSV.