Figure 1.
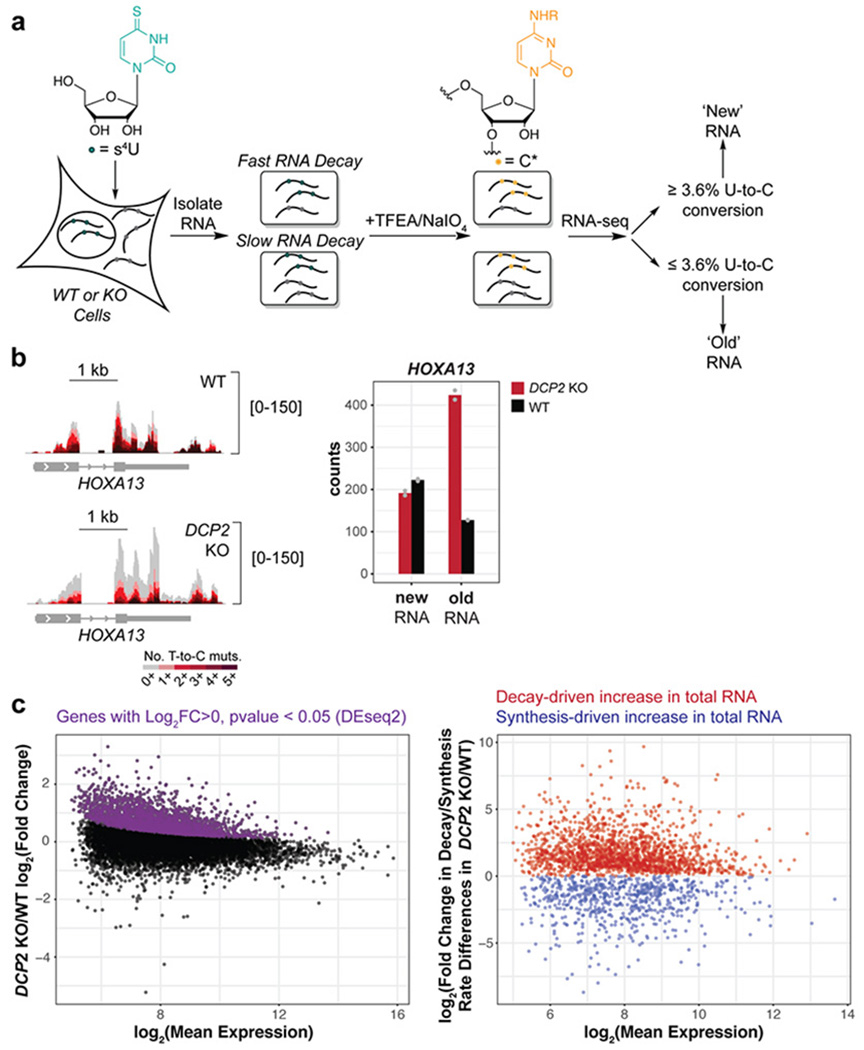
TimeLapse-seq identifies and differentiates global changes in RNA stability and transcription in human cells lacking the cytoplasmic RNA decapping enzyme Dcp2. (a) Experimental scheme of RNA stability measurement by TimeLapse-seq. (b) TimeLapse-seq tracks of HOXA13 in wild type (WT) (top left) and DCP2 knockout (KO) HEK293T cells (bottom left). Reads with no T-to-C mutations represent old RNA and increasing mutational content corresponds to newly synthesized RNA containing recoded s4U incorporated over the 2 h period of metabolic labeling. The bar plot (right) displayed inferred new and old reads from corresponding tracks. (c) Determination of post-transcriptionally stabilized RNAs in DCP2 KO versus WT HEK293T cells. Left, the MA plot indicates significantly upregulated genes (purple) derived from DESeq2 gene-level analysis of total RNAs. Decay- vs. synthesis-driven changes were then separated based on differences in the changes of the estimated rates of degradation and synthesis in DCP2 KO versus WT HEK293T cells (right). Dcp2 targets or substrates are defined as RNAs stabilized in DCP2 KO vs. WT.
