Figure 3.
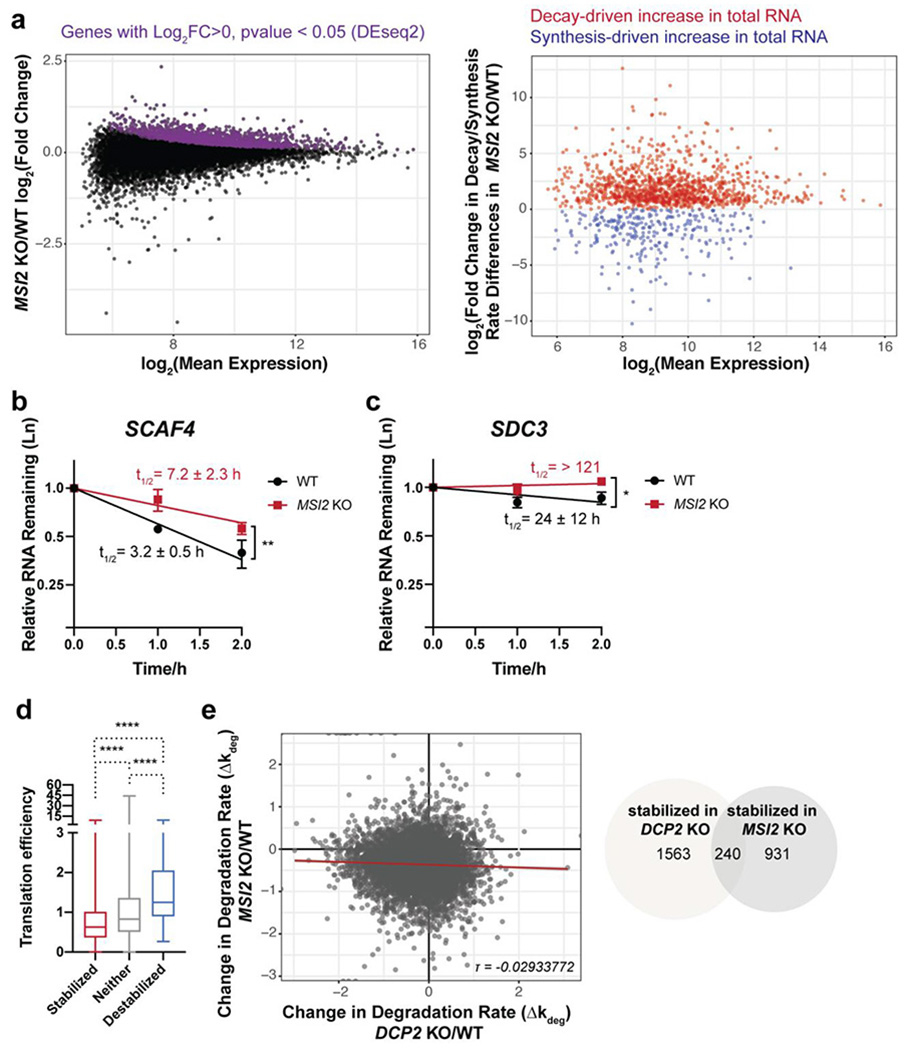
Human Msi2 and Dcp2 regulate distinct subsets of transcripts. (a) TimeLapse-seq analysis of changes in RNA stability and synthesis in MSI2 KO cells. (b-c) qPCR validation in MSI2 KO cells after transcriptional arrest of transcripts stabilized by both MSI2 and DCP2 KO (b), or exclusively by MSI2 KO (c). Error bars represent mean ± s.d. n=3 biological replicates. Half-lives (95% CI) were calculated from linear model, and significance was analyzed by linear regression t-test; *P<0.05; **P < 0.01. (d) Boxplots depicting translation efficiency (data from Sidrauski et al., 2015) for each of the three classes of mRNA stability changes in MSI2 KO versus WT HEK293T cells. Statistical significance is calculated using Mann-Whitney U test; ****P < 0.0001. (e) Msi2 and Dcp2 globally regulate the stability of distinct subsets of RNAs, as shown by low correlation between the changes in estimated RNA degradation rate in each KO relative to WT cells (left, Kendall tau rank correlation coefficient τ=−0.02933772) and low overlap of post-transcriptionally stabilized genes in each KO (right).
