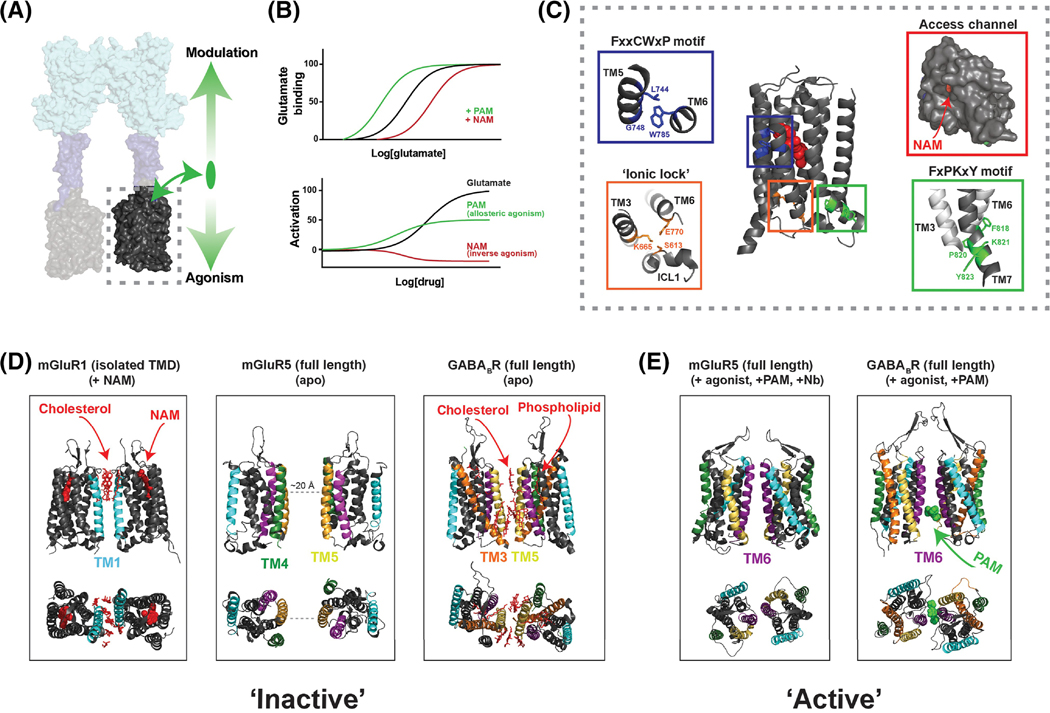
Figure 3, Allosteric modulation and conformational dynamics of class C GPCR transmembrane domains.
A, TMDs (dashed box) respond to agonist-binding in the LBD and mediate G protein activation. TMDs also bind allosteric ligands which can directly alter receptor activation (downward arrow) and modulate the response to orthosteric ligands (upward arrow). B, Theoretical dose-response curves showing the classical effects of allosteric modulators on orthosteric agonist binding (top) and on receptor activation (bottom). Many ligands show properties of both allosteric modulation and agonism or inverse agonism. C, NAM-bound mGluR5 TMD structure (PDB: 4OO9) reveals microswitches similar to those seen in class A structures, including the ionic lock interaction between Lys665 and Glu770 (orange box), the FxxCWxP motif (blue box) which is thought to serve as a “trigger switch” to couple ligand binding to TM6 rearrangement, and the FxPKxY motif (green box) at the intracellular end of TM7 which is thought to stabilize the active conformation. Entrance to the allosteric pocket is restricted by a narrow access channel formed by the helical bundle and extracellular loop 2 (red box). D-E, Structural data showing various inter-TMD dimer interfaces. NAM-bound mGluR1 TMD structures (D, left) revealed an inter-TM1 interface mediated, in part, by cholesterol molecules (red). The cryo-EM structure of mGluR5 showed no direct interface in the apo-state (D, center), but inactive GABABR cryo-EM structures show an interface consisting primarily of TM5 and the cytosolic end of TM3 as well as bound phospholipids and cholesterol (D, right)[23]. The agonist and PAM-bound cryo-EM structure of mGluR5 show an inter-TM6 interface (E, left) [24] that is similar to that seen in agonist and PAM-bound GABABR structures (E, right).