Figure 1. TRAILshort is prevalent within human cancer tissues.
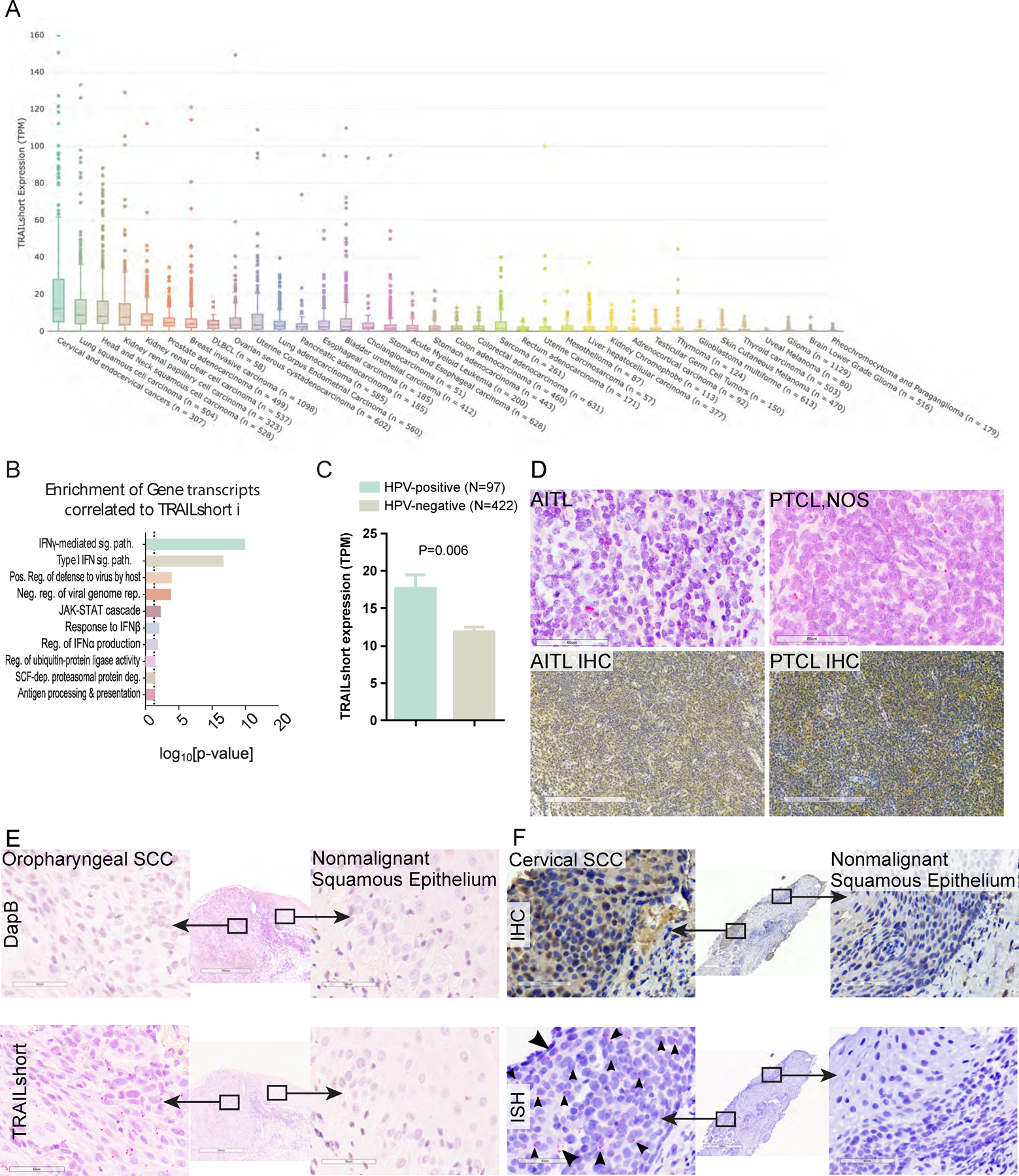
(A) Tissues within the TCGA dataset were queried for the presence of TRAILshort transcripts, and data are expressed as transcripts per million (TPM) according to tissue type. (B) Pearson’s correlation coefficients (PCC) identified 66 gene transcripts co-associated with expression of TRAILshort transcripts, and the functional clustering is shown. (C) TRAILshort transcripts were compared between HPV-positive (N=97) and HPV-negative (N=422) tumors in the TCGA dataset; depicted are mean +/− standard error; P=0.0061 by two-tailed Mann-Whitney test. (D-F) The indicated tissues were stained by immunohistochemistry (IHC) for TRAILshort protein expression (brown staining), or by in situ hybridization (ISH) for TRAILshort mRNA expression (red dots indicated with black arrowheads). (D) Angioimmunoblastic T-cell lymphoma (AITL) and peripheral T-cell lymphoma not otherwise specified (PTCL NOS) stain positive for TRAILshort mRNA ( red dots with black arrowheads). (top panels, RNA ISH, scale bar = 60 µm) as well as by IHC (bottom panels, scale bar = 300 µm). (E) Oropharyngeal squamous cell carcinoma (SCC) tissue was stained by ISH using a negative control probe, DapB top panels, and no signal is detected, yet TRAILshort mRNA is detected by ISH in the malignant tissue indicated by the red dots (highlighted by black arrowheads), but not in the non-malignant adjacent tissue Scale bar = 60 µm for all panels. (F) TRAILshort protein detected by IHC (top panels) or TRAILshort mRNA detected by ISH (bottom panels) in malignant (left panels) or non-malignant (right panels) cervical squamous epithelium.
