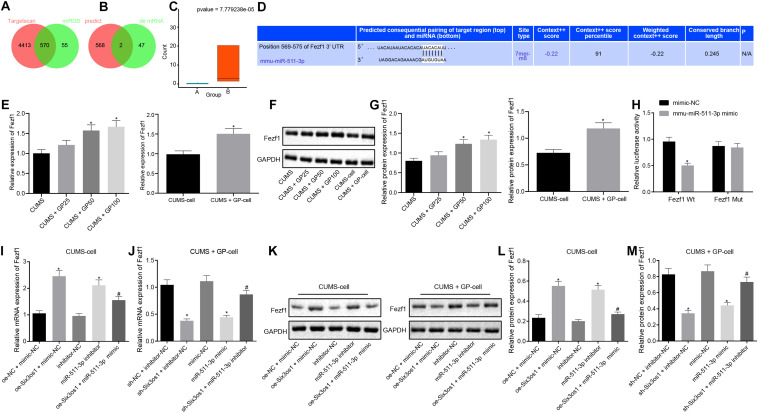
FIGURE 4.
Six3os1 binds to miR-511-3p and upregulates Fezf1. (A) The downstream target genes of miR-511-3p predicted by the TargetScan and miRDB databases; two circles in the figure represent the predicted results of the TargetScan and miRDB databases, with the central part representing the intersection of the predicted results. (B) Intersection of the predicted target genes of miR-511-3p and differentially expressed genes, with the central part representing their intersection. (C) Fezf1 expression detected by RNA-seq data detection, with green boxplot representing CUMS induction and red boxplot representing CUMS induction with GP treatment. The values at upper right were corrected p-values. (D) The targeting relationship between Fezf1 and miR-511-3p predicted by the TargetScan website. (E) Fezf1 mRNA expression in mice (n = 15) and neurons after CUMS and GP treatment detected by RT-qPCR. (F,G) Immunoblots (F) and quantitation of Fezf1 (G) protein expression normalized to GAPDH in mice (n = 15) and neurons after CUMS and GP treatment examined by Western blot analysis. (H) The targeting relationship between Fezf1 and miR-511-3p verified by dual-luciferase reporter gene assay. (I) Fezf1 mRNA expression in CUMS-induced neurons after alteration of Six3os1 and miR-511-3p detected by RT-qPCR. (J) Fezf1 mRNA expression in CUMS-induced neurons with GP treatment neurons after alteration of Six3os1 and miR-511-3p detected by RT-qPCR. (K–M) Immunoblots (K) and quantitation (L,M) of Fezf1 protein expression normalized to GAPDH in CUMS-induced neurons after alteration of Six3os1 and miR-511-3p determined by Western blot analysis. The measurement data were presented as mean ± standard deviation. Data between two groups were compared by independent sample t-test, and data among multiple groups were analyzed by one-way ANOVA, followed by Tukey’s post hoc test. Cell experiment was repeated three times. *p < 0.05 vs. CUMS-induced mice, CUMS-induced neurons, oe-NC + mimic-NC, sh-NC + inhibitor-NC, mimic-NC or inhibitor-NC; #p < 0.05 vs. oe-Six3os1 + mimic-NC or sh-Six3os1 + inhibitor-NC.