Extended Data Fig. 4. ITGB4+ airway progenitors give rise to KRT5+ and SFTPC+ cells that are not from pre-existing Krt5+ and Sftpc+ cells.
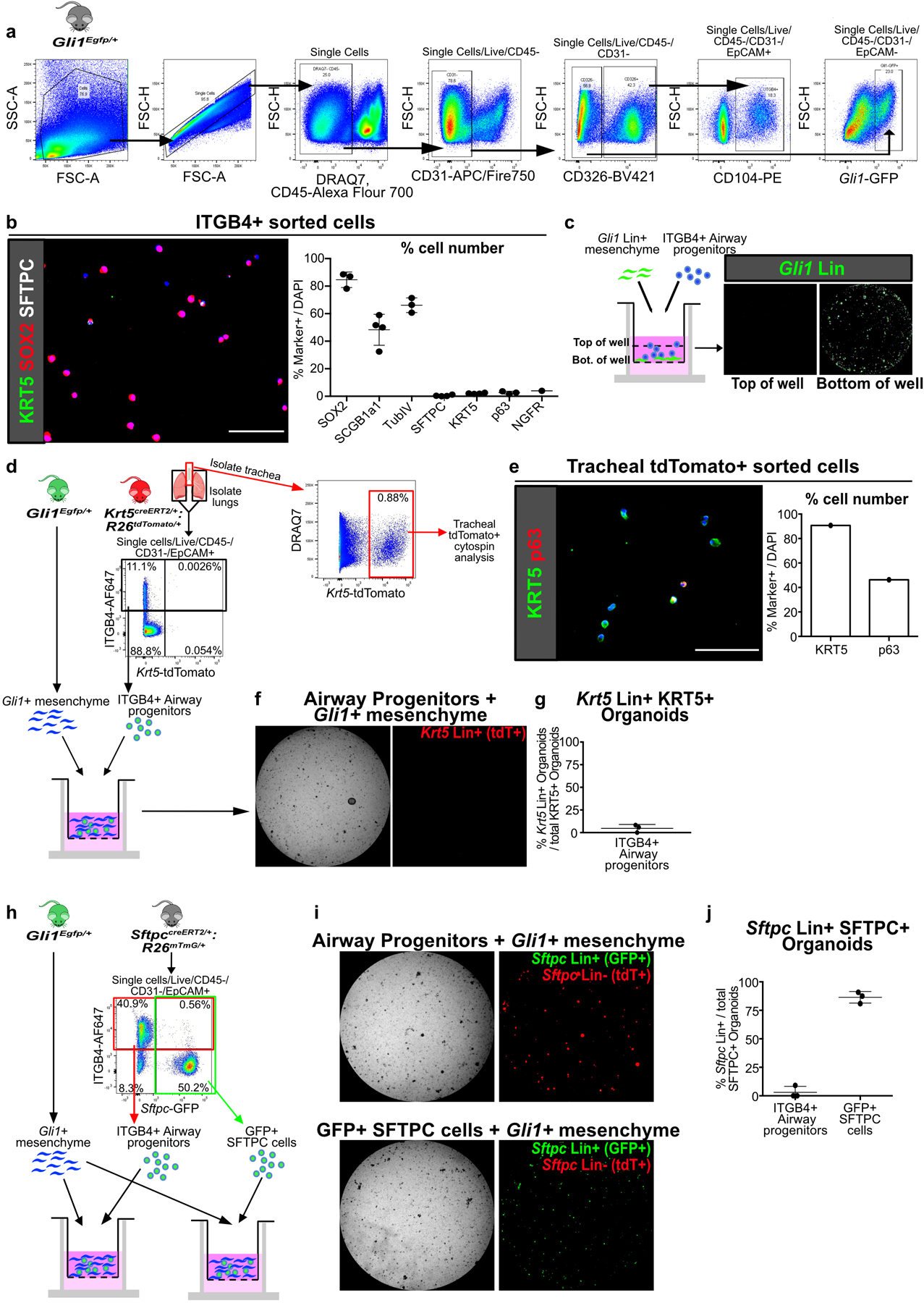
(a) Gating strategy for isolation of ITGB4+ airway progenitors and Gli1+ mesenchymal cells by FACS, presented in figures 2a,e; 3f; 5e; 6f and extended data figures 4d,h; 5h; 8g.
(b) Cytospin of freshly sorted ITGB4+ confirms that majority of cells are SOX2+, SCGB1A1+, and TubIV+, with cells rarely positive for SFTPC, KRT5, p63, or NGFR (n = 1 for NGFR, n = 3 for rest; each data point represents one biological replicate). Data are expressed as mean ± SD.
(c) Z-axis scan shows that Gli1 Lin+ mesenchyme settle to bottom of the Matrigel well.
(d-g) Lineage trace of pre-existing Krt5 Lin+ (tdTomato+) cells followed by isolation of lung for ITGB4+ airway progenitor organoid culture with Gli1+ cells. Tracheal Krt5 Lin+ cells demonstrate expression of basal cell markers KRT5 and p63 (e), while few Krt5 Lin+ cells appear in ITGB4+ cell-derived organoid isolated from the lung of the same animal (f,g). The overwhelming majority of KRT5+ cells in the ITGB4-derived organoids are Krt5 Lin-negative. (n = 3 wells for (f,g); n = 1 sample for (e)). Data are expressed as mean ± SD.
(h-j) Lineage trace of pre-existing Sftpc Lin+ (membrane GFP+) cells followed by isolation of the lung epithelium for organoid culture with Gli1+ cells. ITGB4+ (red box) or Sftpc Lin+ (green box) cells were sorted from the same lung. The overwhelming majority of SFTPC+ organoids in the ITGB4-derived organoids are Sftpc Lin-negative, while most SFTPC+ cells in the Sftpc Lin+ derived organoids are Lin+ (n = 3; each data point represents one well). Data are expressed as mean ± SD. Scale bars, 100 µm.
