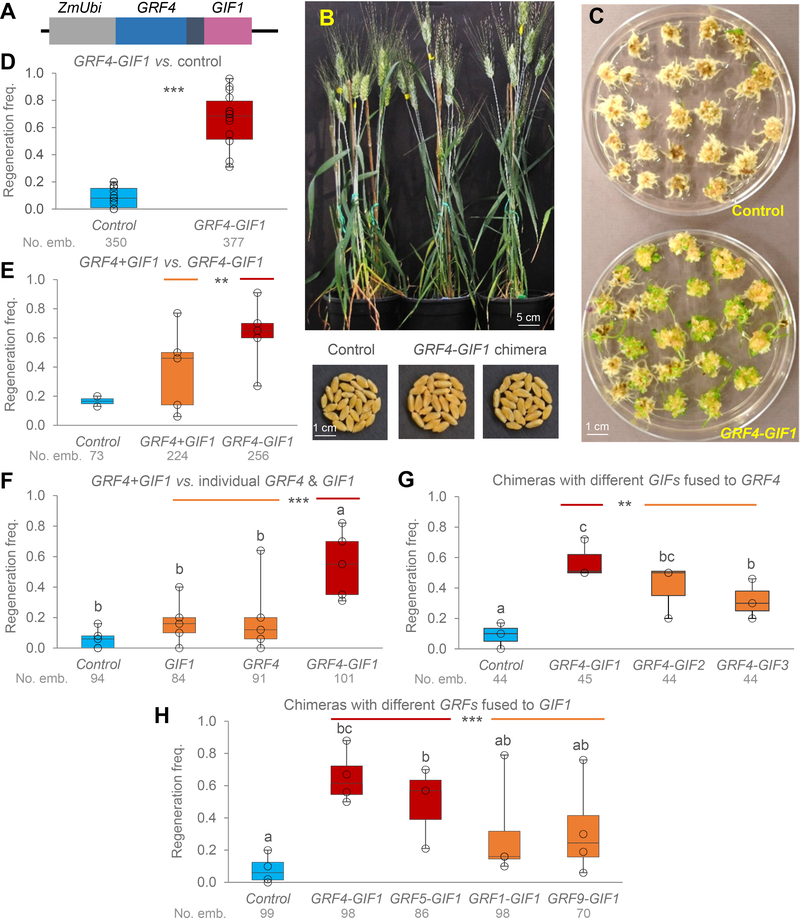
Figure 1. GRF4-GIF1 chimera.
A) Schematic representation of the GRF4 (blue)-GIF1 (pink) chimera. The black region represents a four amino acid spacer. B) The GRF4-GIF1 transgenic wheat plants were normal and fertile. C) Representative transformation showing higher frequency of regenerated shoots during Kronos transformation in the presence of the GRF4-GIF1 chimera than in the control. D-H) Box-plots showing regeneration frequencies of transgenic Kronos plants and their respective controls. The box shows the range from first to third quartiles, and is divided by the median. The whiskers span down to the minimum, and up to the maximum observation. Results from individual experiments are indicated by empty black circles. All experiments include the empty pLC41 vector as control and the wheat GRF4-GIF1 chimera. Numbers below the genotypes are total number of inoculated embryos and different letters above bars indicate significant differences (P < 0.05, Tukey test). D) Control vs. GRF4-GIF1, n= 15 experiments (*** P < 0.001, square root transformation). E) Control, GRF4-GIF1 and vector including GRF4 and GIF1 driven by separate maize UBIQUITIN promoters (GRF4+GIF1), n = 5 experiments (contrast GRF4-GIF1 vs. GRF4+GIF1, ** P = 0.0064, the empty-vector control was included only in two experiments). F) Control, GRF4-GIF1 and vectors including only GIF1 or only GRF4, n = 5 experiments (contrast GRF4-GIF1 vs. combined GRF4 & GIF1 P = 0.0007). G) Control and GRF4 chimeras fused to either GIF1, GIF2 or GIF3, n = 3 experiments (contrast chimeras with GIF1 vs. combined GIF2 and GIF3 ** P = 0.0046). H) Control and chimeras combining different wheat GRF genes fused with GIF1 (n= 4 experiments, except for GRF5 n=3). ** P = 0.0064 in contrast comparing combined GRF4-GIF1 and GRF5-GIF1 chimeras (evolutionary related) with combined GRF1-GIF1 and GRF9-GIF1 chimeras (more distantly related). In all tests, normality of residuals was confirmed by Shapiro-Wilk’s test and homogeneity of variances by Levene’s test (raw-data is available in Supplementary Table 3).