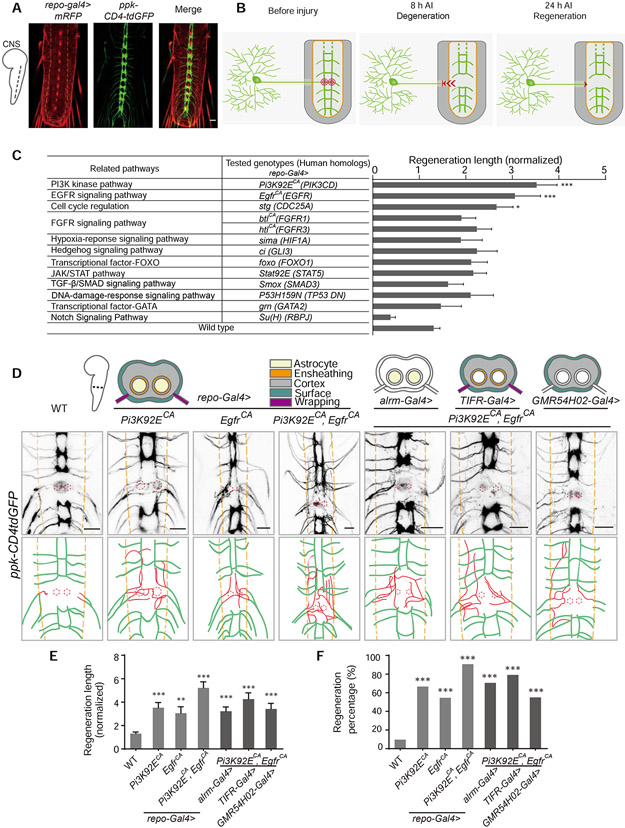
Figure 1. Reprogramming glial cells promotes axon regeneration in the Drosophila CNS.
(A) Images show axon projection of C4da neurons in the VNC of WT Drosophila larvae (ppk-CD4tdGFP; repo-Gal4, UAS-mRFP). Axons are labeled with GFP, glial cells are labeled with RFP, and the neuropil is demarcated by the orange dashed lines.
(B) Schematics of the Drosophila CNS injury model (WT). Left panel shows projection of C4da neuron axons in the larval VNC and the injury sites for one segment (red concentric circles); middle panel shows axons degenerating out of the neuropil (orange region) at 8 h AI; right panel shows injured axons regenerating to the boundary of the neuropil at 24 h AI.
(C) Quantification of normalized regenerated axon length for the screened pathways when manipulated in glial cells, n = 24, 22, 19, 18, 26, 16, 14, 14, 14, 12, 10, 12, 18, 62 lesioned segments from 12, 11, 10, 9, 13, 8, 7, 7, 7, 6, 5, 6, 9, 31 larvae respectively for each genotype, WT (ppk-CD4tdGFP; repo-Gal4, UAS-mRFP) as control, one-way ANOVA with Dunnett’s test. See also Table S1.
(D) Axon regrowth of C4da neurons induced by expressing Pi3K92ECA and EgfrCA individually or together in glia under the control of repo-Gal4, alrm-gal4, TIFR-Gal4 or GMR54H02-Gal4 at 24 h AI on segment A3 in the VNC. Schematics above the images show glia cohorts labeled by different promoters; diagrams below the images show regenerated axons (red) in the neuropil (demarcated by the orange dashed lines); the red dotted circles show injury sites.
(E) Quantification of normalized regenerated axon length for genotypes shown in (D), n = 62, 24, 22, 32, 34, 24, 20 lesioned segments from 31, 12, 11, 16, 17, 12, 10 larvae respectively for each genotype, one-way ANOVA with Dunnett’s test.
(F) Quantification of regeneration percentage for genotypes shown in (D), Fisher’s exact test.
*P < 0.05, **P < 0.01 and ***P < 0.001. Scale bars, 20 μm. Data are expressed as mean ± s.e.m. CA, constitutively active; DN, dominant negative. See also Figure S1.