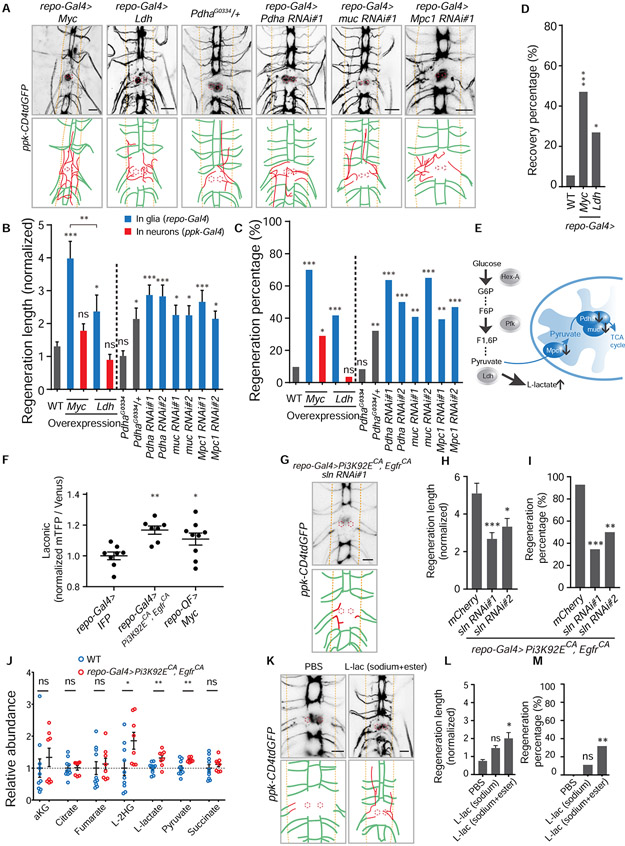
Figure 4. Elevated glycolysis in glia is sufficient to promote axon regeneration in the CNS via glial L-lactate efflux.
(A) Axon regrowth of C4da neurons in flies with glial Myc expression, glial Ldh expression, Pdha mutation, glial Pdha RNAi, glial muc RNAi and glial Mpc1 RNAi at 24 h AI on segment A3 in the VNC. Diagrams below the images show regenerated axons (red) in the neuropil (demarcated by the orange dashed lines), and the red dotted circles show injury sites.
(B) Quantification of normalized regenerated axon length for the genotypes of two separated groups. Left: WT, Myc expression in glia or C4da neurons, Ldh expression in glia or C4da neurons; right (WT as control): Pdha heterozygous and homozygous mutants, glial Pdha RNAis, glial muc RNAis and glial Mpc1 RNAis, with two RNAis for each gene knockdown, n = 62, 20, 31, 24, 28, 24, 28, 22, 28, 27, 20, 28, 32 lesioned segments from 31, 10, 16, 12, 14, 12, 14, 11, 14, 14, 10, 14, 16 larvae respectively for each genotype, one-way ANOVA with Holm-Sidak's test.
(C) Quantification of regeneration percentage for genotypes in (B), Fisher’s exact test.
(D) Functional recovery percentage of larvae (WT as control) in which glial cells express Myc or Ldh, n = 36, 17, 26 larvae for each genotype, Fisher’s exact test.
(E) Schematic shows blocking genes which functionally connect glycolysis and the TCA cycle will lead to increase of the end-product of glycolysis, L-lactate.
(F) Lactate level revealed by Laconic driven by repo-Gal4. Quantifications of fluorescence intensity ratio of mTFP/Venus from glial cells in the VNC, glial IFP expression as control (UAS-IFP; repo-Gal4, UAS-Laconic), n = 8, 7, 9 VNCs for each genotype, one-way ANOVA with Dunnett’s test.
(G) Image and diagram showing C4da neuron axon regrowth of flies with glial sln RNAi in the background of repo-Gal4>Pi3K92ECA, EgfrCA at 24 h AI on segment A3 in the VNC.
(H) Quantification of normalized regenerated axon length for the two glial sln RNAis in the background of repo-Gal4>Pi3K92ECA, EgfrCA, glial mCherry expression as control (UAS-mCherry; repo-Gal4, UAS-Pi3K92ECA, UAS-EgfrCA; ppk-CD4tdGFP), n = 28, 26, 18 from 14, 13, 9 larvae respectively lesioned segments for each genotype, one-way ANOVA with Dunnett’s test.
(I) Quantification of regeneration percentage for genotypes in (H), Fisher’s exact test.
(J) Metabolites level in the hemolymph collected from larvae of repo-Gal4>Pi3K92ECA, EgfrCA (n = 8) normalized to those in WT (n = 10), unpaired two-tailed Student's t-test for each metabolite.
(K) Axon regrowth of C4da neurons at 24 h AI on segment A3 in the VNC of WT flies injected with PBS, L-lactate (final cone., ~100 mM sodium L-lactate + 15 mM ethyl L-lactate) right after injury.
(L) Quantification of normalized regenerated axon length for WT flies injected with PBS, sodium L-lactate only (final con., ~150 mM) and L-lactate (final conc., ~100 mM sodium L-lactate + 15 mM ethyl L-lactate) right after injury, n = 22, 18, 22 lesioned segments from 11, 9, 11 larvae respectively for each group, one-way ANOVA with Dunnett’s test.
(M) Quantification of regeneration percentage for treatment groups in (L), Fisher’s exact test.
*P < 0.05, **P < 0.01, ***P < 0.001 and ns, not significant. Scale bars, 20 μm. Data are expressed as mean ± s.e.m. CA, constitutively active. See also Figure S4.