Figure 3.
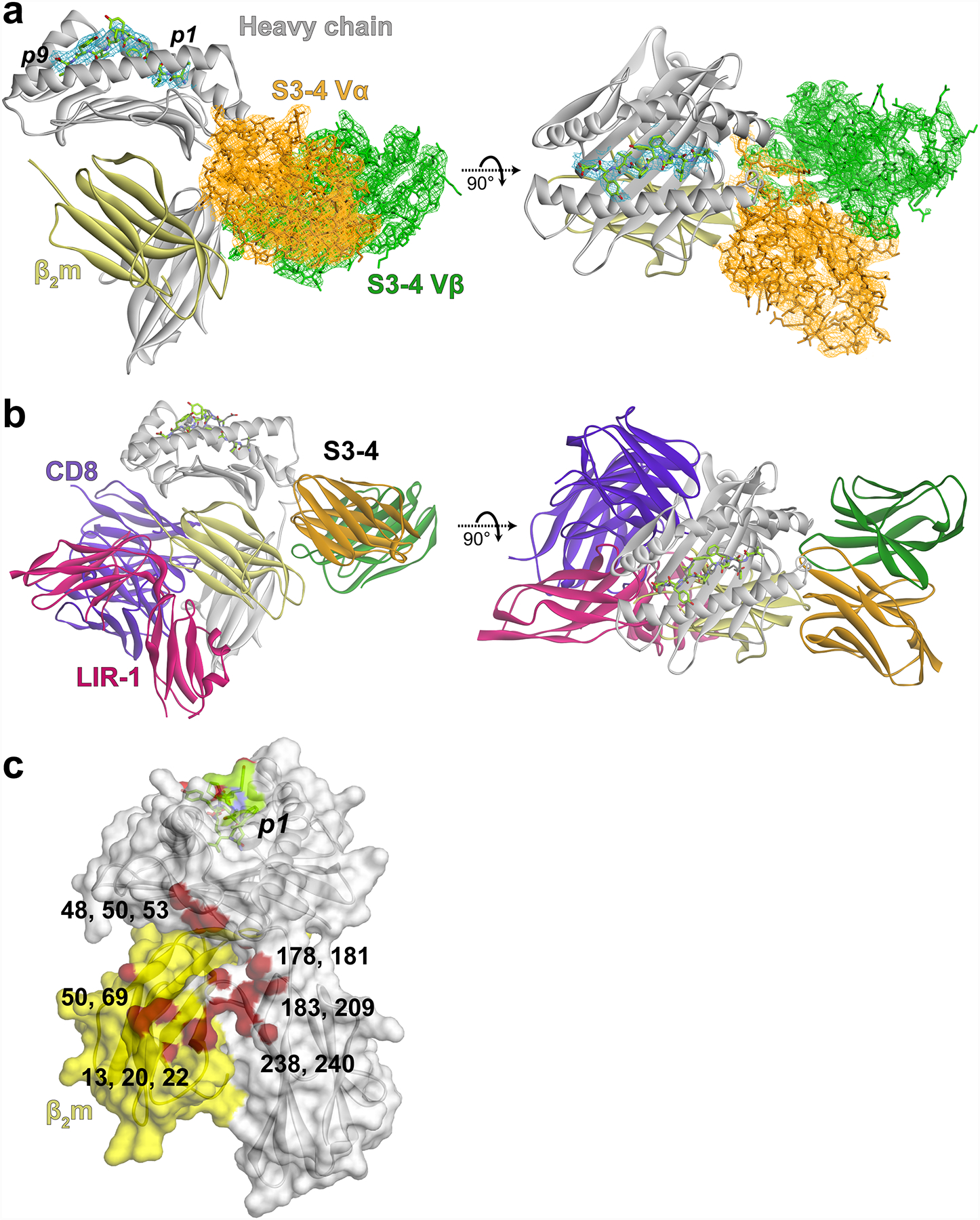
New and altered molecular interactions in the interface between S3–4 and HLA-A2. A) The new arginine at position 100 in CDR3α extends into a cleft on the side of HLA-A2, interacting with Thr178, Arg181, and Tyr209. These interactions are facilitated by replacement of the bulky Trp101α with serine. The S3–4 Vα domain is orange, the Vβ domain green, and the HLA-A2 heavy chain is gray. The wild-type A6 TCR, modeled by superimposing its variable domains onto those of S3–4, is colored light blue. This color scheme is maintained throughout the figure. B) Mutation of Asp26 in CDR1α to isoleucine allows hydrophobic packing with Pro50 of HLA-A2 and removes electrostatic repulsion that would exist between the original aspartic acid and Glu53 of HLA-A2, which interacts with the backbone of Arg27 and Ser28 in CDR1α. C) Mutation of Leu98 in CDR3β to alanine and an associated conformational change allows the loop to avoid steric clashes with Asp183 and Lys186 of HLA-A2. D) In addition to forming new interactions and allowing the TCR to avoid steric and electrostatic clashes, the mutations in S3–4 would destabilize the normal TCR binding mode. Replacement of Gln30 in CDR1α, for example, would remove key hydrogen bonds between the TCR and the peptide backbone. Replacement of Ser100 in CDR3α with arginine would result in clashes with the center of the peptide.
