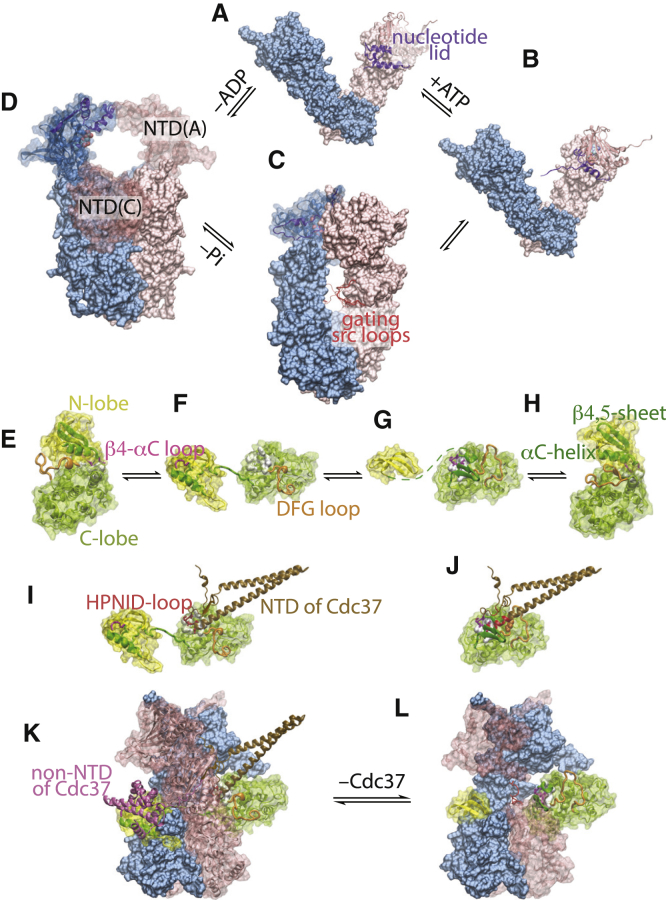
Figure 1.
Structural basis for my model of nonequilibrium activation of client kinases by Hsp90. The underlined texts below are Protein Data Bank IDs and chain IDs. The Hsp90 homodimer goes through four states in A to D. (A) Hsp90open: nucleotide free and open; 2IOQ. (B) : ATP bound and open; 2IOQ but with the NTD of one chain replaced with the ATP-bound NTD from the structure 2CG9. ATP binding induces a structural change in the nucleotide lid in NTD. (C) : ATP bound and closed; 2CG9. The hole between the two MDs in the closed Hsp90 dimer is gated by a flexible, often disordered src loop (the src loops from 2CG9, 5FWM, 2IOP, and 4IYN are superimposed). (D) : ADP-bound and compact; 2IOP. The ADP-bound NTD may have a number of orientations with respect to MD; e.g., the NTD of chain A may move to the position occupied by the NTD of chain C in the crystal structure, resulting in a compact conformation consistent with electron microscopy images (12) but clashing with the bound kinase client. In (B)–(D), the nucleotides are shown in spheres. The kinase client may transition through four states in (E) to (H). (E) The inactive conformation: i; 1HCL:A. (F) The deactivating conformation: d; 5FWM:K. (G) The hypothetical maturing conformation: m. (H) The active conformation: a; 1FIN:A. The deactivating structure is of the client kinase Cdk4 in the Hsp90-Cdc37-Cdk4 complex; the inactive and active structures are of Cdk2. (I) Complex between Cdc37 and Cdk4 in the d-state (5FWM:K + E) is shown. (J) Steric clash prevents Cdc37 from binding to Cdk4 in the m-state. Three residues (in spheres) in the DFG loop—in addition to perhaps the β4-αC loop—would collide with Cdc37, whose clashing residues are highlighted in red. (K) The ternary complex of Hsp90-Cdc37-Cdk4 (5FWM), in which the client kinase is in the d-state, is shown. (L) The hypothetical complex of the closed Hsp90 dimer with the client kinase in the m-state is shown. To see this figure in color, go online.