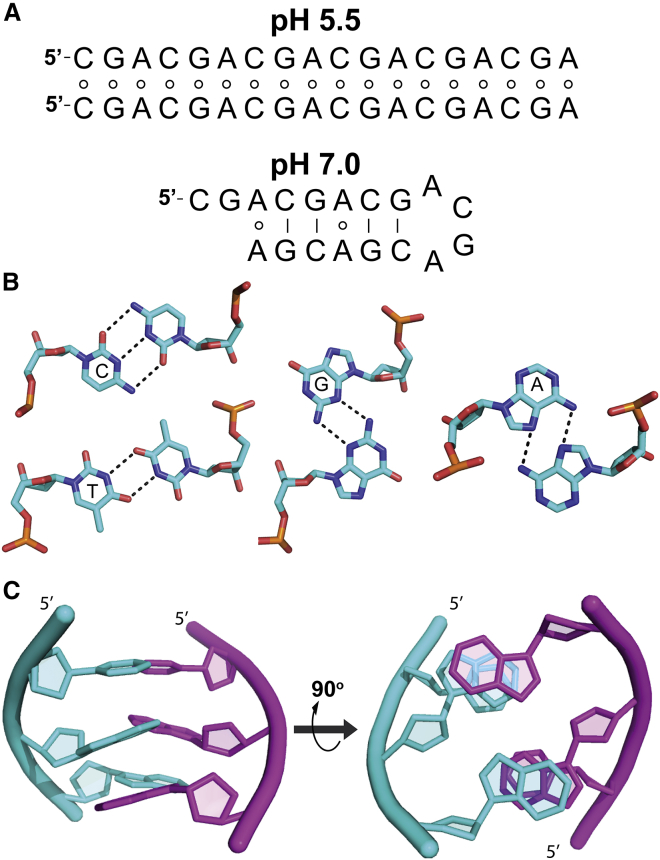
Figure 1.
d(CGA) triplet motif. (A) (CGA)6 is predicted to form a parallel-stranded homoduplex at pH 5.5 and an antiparallel hairpin at pH 7.0, based on NMR and CD analysis (25, 26, 27, 28). Circles indicate noncanonical base pairing, and solid lines represent canonical base pairing. It is unclear whether the antiparallel hairpin anticipated at pH 7.0 forms with (shown) or without (not shown) A-A homo-base pairs (25). (B) Shown are the following homo-base pairing interactions in the parallel-stranded form of the d(YGA) motif: protonation of cytosine N3 allows formation of a hemiprotonated C-CH+ homo-base pair, T-T homo-base pair, G-G minor groove edge homo-base pair, and A-A Hoogsteen-face homo-base pair. (C) A d(CGA) triplet forming a parallel-stranded homoduplex, shown from the side and also rotated 90° so the 5′ ends go into the page, to show interstrand GA stacking interactions. Schematics in (B) and (C) used Protein Data Bank structures PDB: 4RIM and PDB: 5CV2. To see this figure in color, go online.