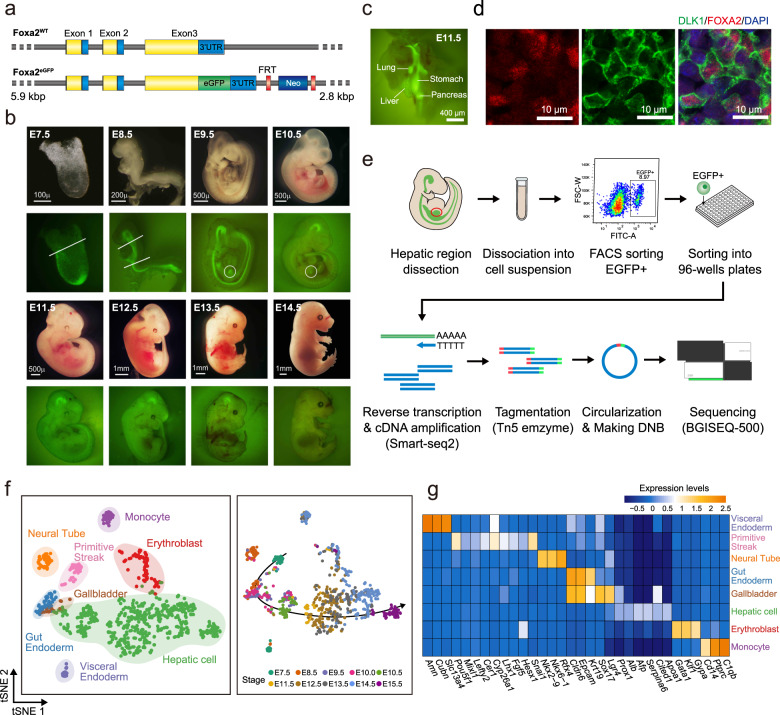
Fig. 1. Single-cell full-length RNA-seq to analyze liver development during E7.5–E15.5 by using a Foxa2 eGFP mouse model.
a The vector design for Foxa2eGFP mouse. eGFP is linked to the third exon of Foxa2. CDS coding sequence, 3′UTR 3′ untranslated region, FRT flippase recognition target, Neo Neomycin. b eGFP-labeled mouse embryos from E7.5 to E14.5. The endoderm, neural system, and endoderm-derived organs, including liver expressed eGFP. The general dissection strategies are shown (white lines or circles). Scale bars, 100 μm for E7.5; 200 μm for E8.5; 500 μm for E9.5–E11.5; 1 mm for E12.5–E14.5. c Precise dissection for the endodermal organs at E11.5. The liver, lung, stomach, and pancreas are shown. Scale bars, 400 μm. d Immunofluorescence analysis of paraffin-sectioned mouse embryo at E14.5, showing co-expression of FOXA2 (red), DLK1 (green), and DAPI (blue) in hepatoblasts. Scale bar, 10 µm. e The workflow of single-cell full-length RNA sequencing of Foxa2eGFP+ cells. f t-SNE visualization of embryonic cells from E7.5 to E15.5, colored by cell populations and embryonic stages. The left panel shows the cell identity of each cell cluster, and the right panel shows the embryonic stage of every single cell. The developmental route (shown by the arrow in the right panel) by t-SNE agrees with the embryonic stages of single cells. g Expression of selected marker genes for the identified cell clusters. The averaged gene expression level of marker genes are shown by different colors.