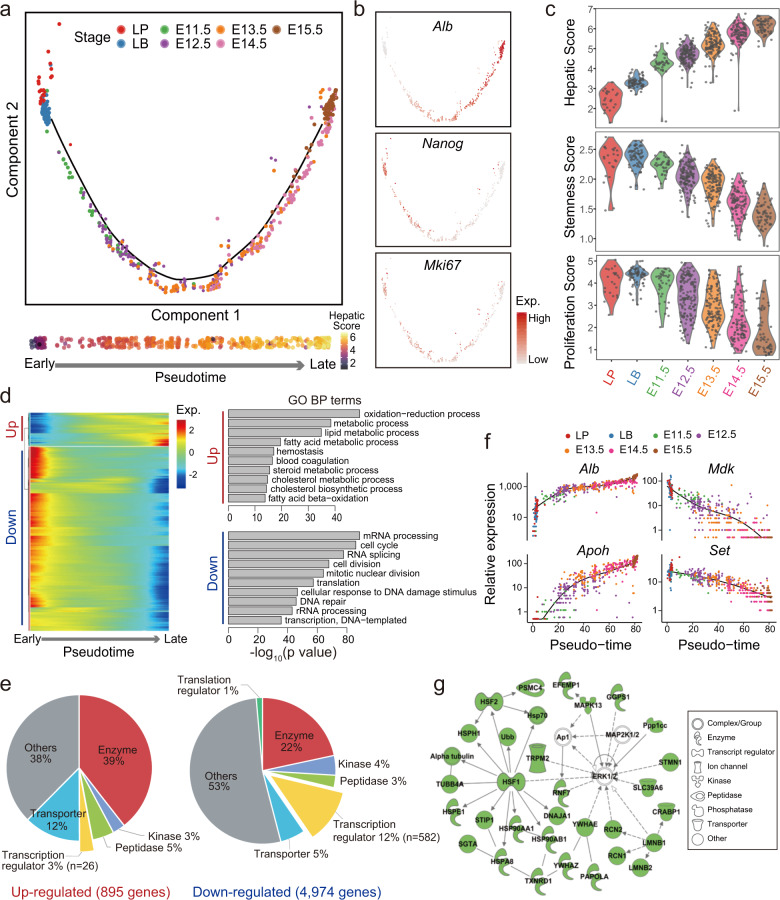
Fig. 4. Dynamic gene expression during hepatoblast maturation into hepatocytes.
a A trajectory of hepatic development was determined during E9.5–E15.5 by Monocle analysis. No branches on the trajectory were found, and the predicted pseudotime of the trajectory agreed with the gestation day. b The expressions of Alb, Nanog, Mki67 were shown in the hepatic trajectory. c The metabolic function of hepatoblasts/hepatocytes increased while the cell pluripotency and the proliferation rate decreased during liver development based on “hepatic score”, “stemness score”, and “proliferation score” from E9.5 to E15.5. d Dynamic regulation of gene expression during hepatoblast maturation and the respective functions of up/downregulated genes were identified. e The composition of upregulated or downregulated genes during E9.5–E15.5 were shown. f Genes for the metabolic function of the liver (such as Alb and Apoh) from E9.5 to E15.5 were upregulated, while the downregulated genes were enriched in cell cycles, RNA splicing, cell division, and translation (such as Mdk and Set). g The network of genes that were downregulated during the hepatoblasts maturation. Genes labeled by green were downregulated, while genes labeled by white were not detected.