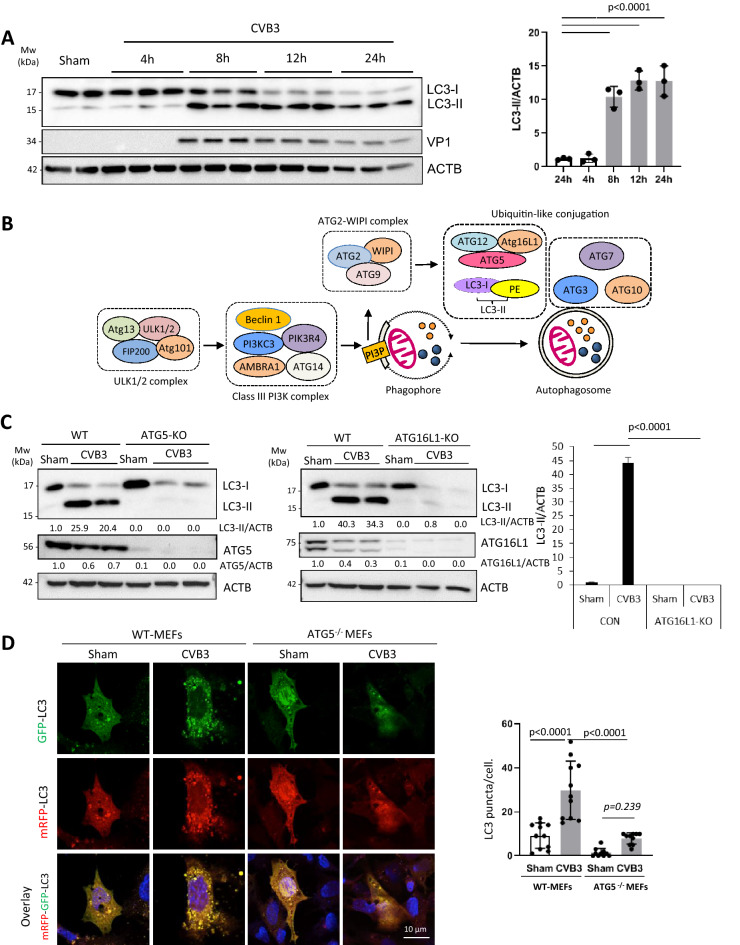
Figure 1.
CVB3-induced LC3 puncta and lipidation are dependent on ATG5 and ATG16L1. (A) HEK293A were either sham-infected with PBS or infected with CVB3 for indicated time-points. Western blotting was performed to detect LC3 lipidation and ACTB expression (loading control). Levels of LC3-II were quantified by densitometric analysis, normalized to ACTB and presented as fold changes in the right panel (mean ± SD, n = 3, analyzed by one-way ANOVA with Tukey’s post-test). (B) Schematic diagram of canonical autophagy pathway induced by amino acid deprivation. (C) ATG5- or ATG16L1-knockout (KO) HEK293A cells generated via CRISPR-Cas9 editing, or wildtype (WT)-HEK293A cells were either sham- or CVB3-infected for 16 h. Cell lysates were harvested and probed for LC3, ATG5, and ATG16L1 by Western blotting. Densitometric analysis was carried out as above (the first lane is arbitrarily set a value of 1) and the results are presented under each blot. (D) WT- or Atg5−/− MEFs transfected with mRFP–GFP–LC3 construct were sham- or CVB3-infected for 8 h. GFP and RFP signal was captured by confocal microscopy. Mean LC3 puncta per cell was quantified (n ≥ 10 cells) and analyzed by one-way ANOVA with Tukey post-test (right).