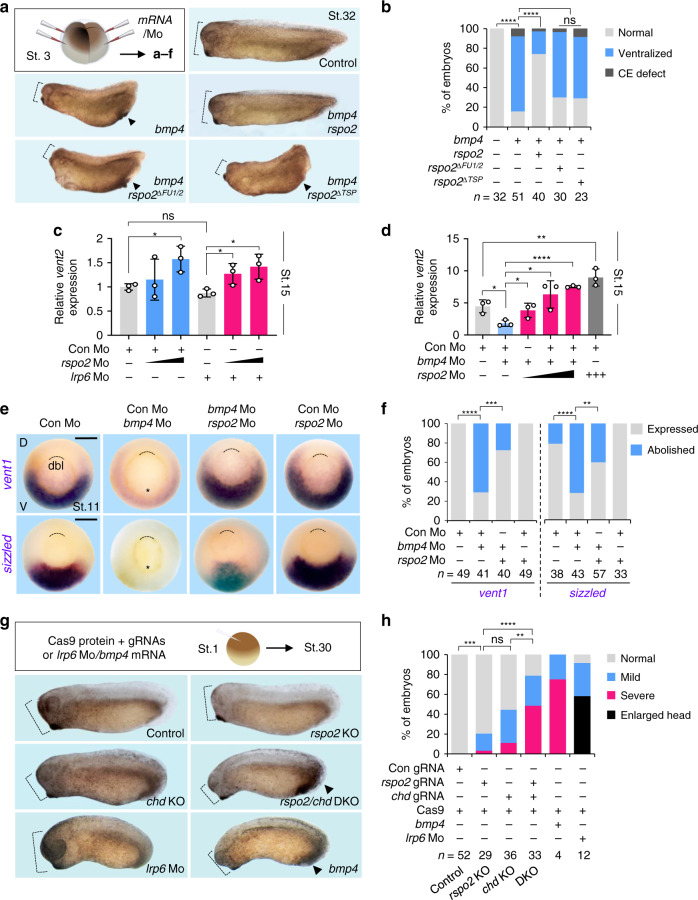
Fig. 2. Rspo2 inhibits BMP4 signaling in Xenopus dorsoventral embryonic patterning.
a Microinjection strategy for a–f, and representative phenotypes of Xenopus laevis tadpoles (St. 32) injected with the indicated mRNAs radially at 4-cell stage. Dashed lines, head size. Arrowheads, enlarged ventral structure. b Quantification of embryonic phenotypes shown in a. ‘Ventralized’ represents embryos with both small head and enlarged ventral structure, reminiscent of BMP hyperactivation. ‘CE defect’ refers to embryos with convergent extension (gastrulation) defects, unrelated to BMP signaling. Note that rspo2 mRNA dosage used in a was below those that cause gastrulation defects. n = number of embryos. c, d BMP-(vent2) reporter assays with Xenopus laevis neurulae (St.15) injected with reporter plasmids and the indicated Mo at 4-cell stage. n = biologically independent samples and data are displayed as means ± SD. ns, not significant. *P < 0.1, **P < 0.01, ****P < 0.0001 from two-tailed unpaired t-test. e In situ hybridization of vent1 and sizzled in Xenopus laevis gastrulae (St.11, dorsal to the top, vegetal view) injected as indicated. D, dorsal, V, ventral. Asterisk, abolishment of the expression. Dashed line, dorsal blastopore lip (dbl). Scale bar, 0.5 mm. f Quantification of embryonic phenotypes shown in (e). ‘Expressed’, normal, increased or reappearance of vent1/sizzled expression. ‘Abolished’, complete absence of vent1/sizzled expression. Data are pooled from two independent experiments. n = number of embryos. g Microinjection strategy and representative phenotypes of Xenopus tropicalis tadpole (St.30) Crispants and tadpoles (St.30) injected with bmp4 mRNA or lrp6 Mo. At 1-cell stage, Cas9 protein with guide RNA (gRNA) targeting rspo2 or chd, or both gRNAs were injected animally. Dashed lines, head size. Arrowheads, enlarged ventral structure. h Quantification embryonic phenotypes shown in g. ‘Severe’ showed small head, enlarged ventral tissues and short body axis. ‘Mild’ showed one or two of the defects described above. ‘Normal’ showed no visible differences to the uninjected control. n = number of embryos. ns, not significant. **P < 0.01, ***P < 0.001, ****P < 0.0001 from two-tailed χ2 test comparing normal versus ventralized phenotypes (b), two-tailed χ2 test comparing expressed versus abolished (f), or two-tailed χ2 test comparing normal versus severe and mild defects h.