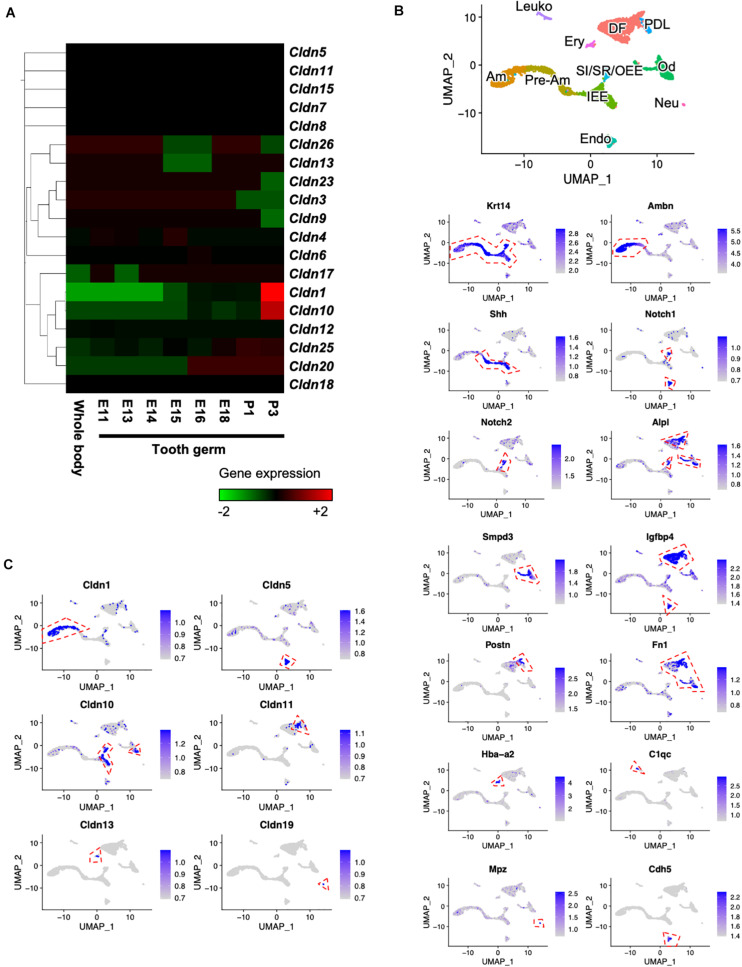
FIGURE 1.
Expression patterns of claudin family members during tooth development. (A) Heatmap of Cap Analysis Gene Expression (CAGE)-based claudin genes expression in embryonic (E) day E11 mouse whole body and tooth germs at various developmental stages. (B) Differential expression analysis of cell-type marker genes using Uniform Manifold Approximation and Projection (UMAP) visualization of single-cell datasets from postnatal (P) day P7 Krt14-RFP mouse incisors. Krt14, epithelium marker; Ambn, ameloblast marker; Shh, IEE and pre-ameloblast marker; Notch1 and Alpl, SI markers; Notch2, SR and OEE marker; Smpd3, odontoblast marker; Igfbp4, dental follicle and Postn, periodontal ligament markers; Fn1, mesenchyme marker; Hba-a2, erythrocyte marker; C1qc, leukocyte marker; Mpz, neural cell marker; Cdh5, endothelial cell marker. IEE, inner enamel epithelium; Pre-Am, pre-ameloblast; Am, ameloblast; SI/SR/OEE, stratum intermedium, stellate reticulum, and outer enamel epithelium; DF, dental follicle; Od, odontoblast; PDL, periodontal ligament; Ery, erythrocyte; Leuko, leukocyte; Endo, endothelium; Neu, neural cell. Red dashed lines indicate the cell cluster that highly expressed marker genes. (C) Differential gene expression analysis of claudin family members detected in single-cell datasets from P7 Krt14-RFP incisors. Red dashed lines indicate the cell cluster that highly expressed marker genes.