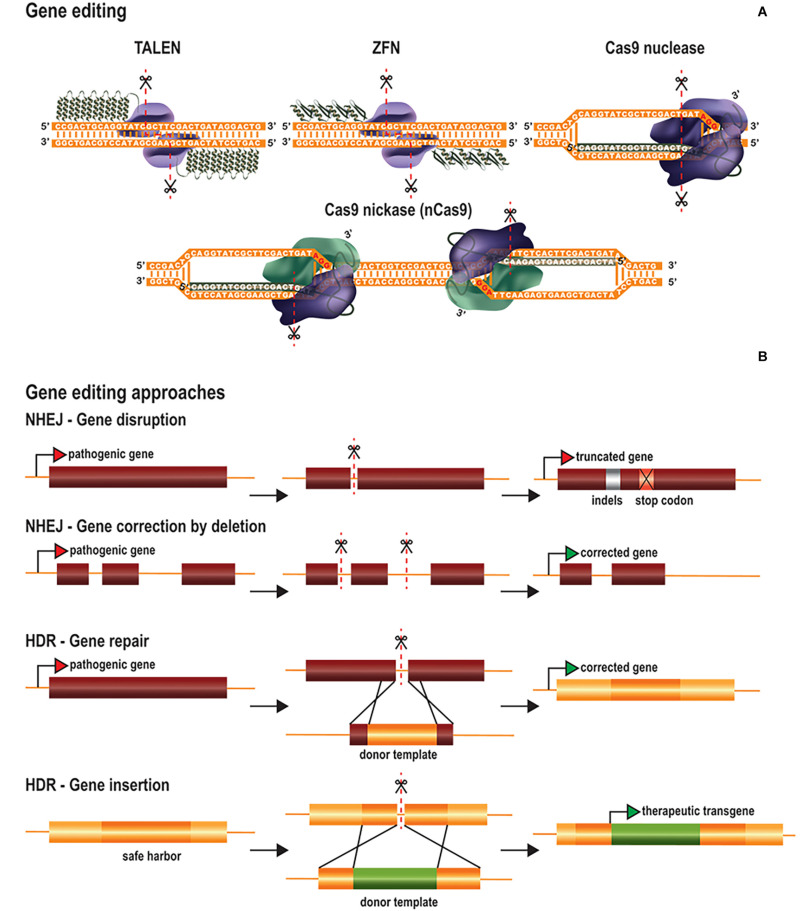
FIGURE 1.
Gene editing tools and therapeutic approaches. (A) Gene editing tools are based on TALEs, ZFs and CRISPR/Cas platforms. Site-specific TALENs and ZFNs consist of two modules of TALEs and ZFs fused to the FokI nuclease. Both modules recognize adjacent sequences in opposite strands to promote the dimerization of FokI and sequence cleavage in a staggered fashion. In contrast, CRISPR/Cas systems hold intrinsic nuclease activity. Cas nucleases or Cas nickases are explored to produce either DSBs or SSBs in the targeted sequence, respectively. Alternatively, paired nickases targeting adjacent sequences in opposite strands generate staggered DSBs. (B) Gene editing therapeutic approaches rely on the intrinsic DNA repair mechanisms NHEJ and HDR after generation of DSBs. Gene disruption by NHEJ involves the introduction of indels after generation of DSBs at the coding region of a pathogenic gene, resulting in the formation of a premature stop codon. Gene correction by NHEJ implicates the targeting of the non-coding region of a pathogenic gene. It includes the removal of deleterious exons by the simultaneous cleavage in both upstream and downstream intronic regions and/or disruption of splicing regulation sites. Both gene repair and gene insertion by HDR involve the use of donor templates containing intended sequences flanked by homology arms. In the first case, the template is targeted to the pathogenic gene and contains the corrected sequence allowing gene restoration. In contrast, gene insertion by HDR targets safe harbor locations in the genome to introduce therapeutic transgene expression cassettes.