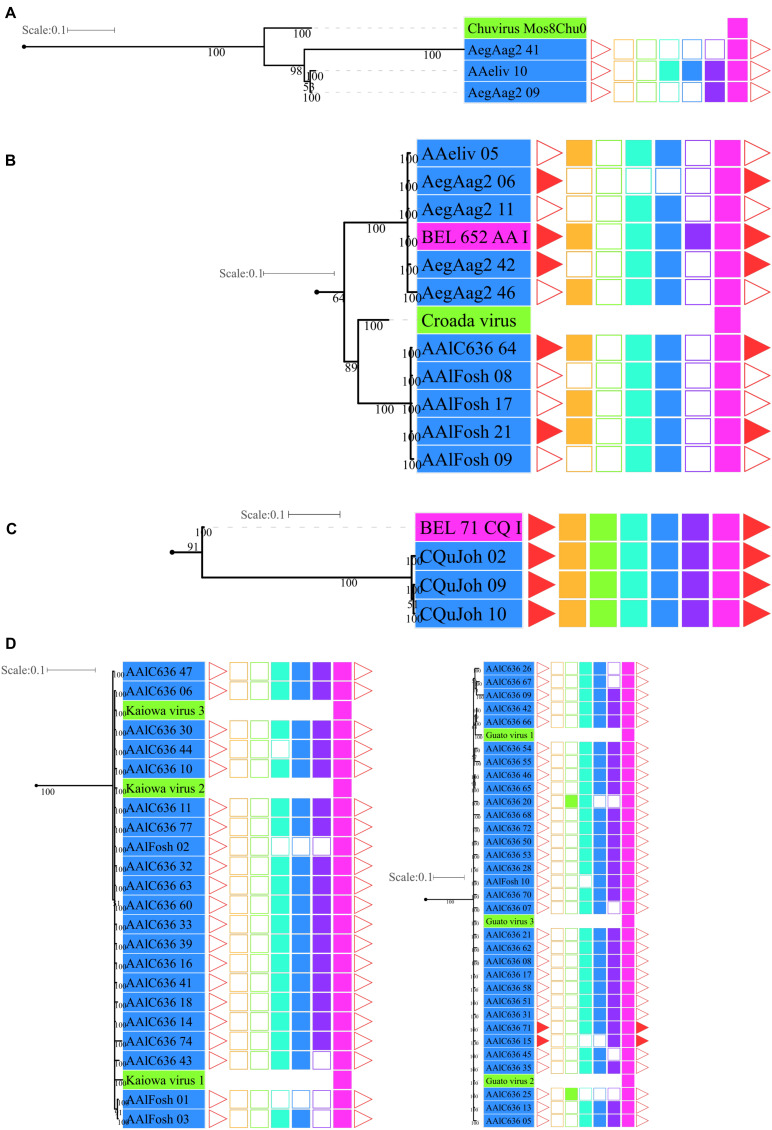
FIGURE 5.
Phylogenetic clades showing particular clustering of some sequences from Figure 4. (A) Basal clade with bona fide chuvirus, chuvirus solo-glycoprotein and BEL-Pao derived elements associated with chuvirus glycoprotein; (B) clades with a number of highly similar glycoproteins found associated with BEL-Pao elements; (C) clade with complete BEL-Pao Retrotroelements (Anakin); (D) clades with BEL-Pao derived elements associated with chuvirus glycoprotein and potential chuvirus sequences described by Lara Pinto et al. (2017). Sequences identified as EVEs derived from Chuviridae family, blue and red color range labels represent EVEs identified in Culicinae and Anophelinae subfamilies, respectively, retrotransposons of BEL-Pao superfamily available on RepBase and that showed similarity with chuviruses glycoproteins, range labels colored with purple; Chuviruses identified in China by Shi et al. (2016), range labels colored with light orange; Sequences described in Brazil as chuviruses by Lara Pinto et al. (2017), range labels colored with light green; Chuvirus available on NCBI (access KX924631.1), range label colored with dark orange; LTR, Long Terminal Repeat—red triangle; GAG, Capsid protein—orange square; PR, Protease—light green square; RT, Reverse Transcriptase—light blue square; RH, RNAse H—blue square; Integrase—purple square and GLY, Glycoprotein—pink square.