FIG 1.
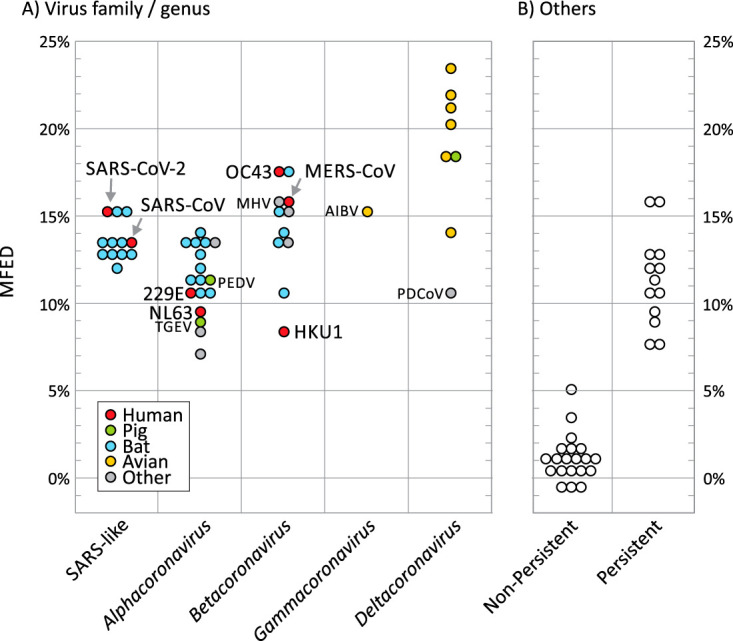
RNA structure prediction in coronaviruses and previously characterized persistent/nonpersistent positive-strand RNA viruses. RNA structure formation was predicted by comparison of minimum folding energies of virus native sequences with those of shuffled controls (MFED value on the y axis). (A) Data points represent MFEDs for type member of each currently classified coronavirus species (listed in Table S1 in the supplemental material) and a separate category for SARS-CoV-2, SARS-CoV, and a range of SARS-like viruses infecting bats (sarbecoviruses). Human viruses and widely investigated coronaviruses infecting other species are labeled. AIBV, avian infectious bronchitis virus; MHV, mouse hepatitis virus; PDCoV, porcine deltacoronavirus; PEDV, porcine epidemic diarrhea virus; TGEV, transmissible gastroenteritis virus. (B) MFED values of previously analyzed positive-strand mammalian viruses from a previous study and that reported the association between RNA structure and persistence (19).
