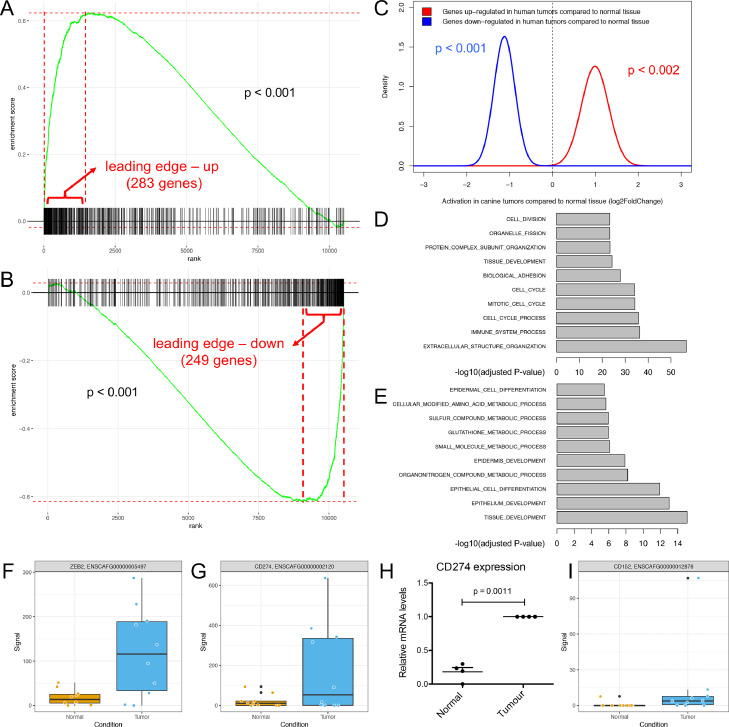
Figure 2.
COSCC and HNSCC display a high grade of homology. (A and B) Competitive gene set testing to compare COSCC to HNSCC. GSEA-like running sum statistic depicting the location of (A) up-regulated and (B) down-regulated genes in HNSCC on a ranked list of genes in COSCC compared to normal. Permutation P-values were calculated by fgsea package. (C) Self-contained gene set testing (QuSAGE method) to assess the average differential expression of HNSCC gene sets (i.e., up- and down-regulated genes in HNSCC) in COSCC. X-axis demonstrates mean fold change expression in tumor compared to normal in COSCC (HNSCC data obtained from GSE62944). Y-axis indicates the distribution of fold change expression within each set. P-values were calculated by comparing mean fold change to fold change of 1 using Welch's t-test. (D-E) Hypergeometric P-values indicating enrichment of Gene Ontology biological processes (GObp) among the leading edge up-regulated (D) and down-regulated (E) genes as revealed by competitive gene testing in panels A and B. (F) Levels of ZEB2 in normal epithelium and tumor cells as detected by RNAseq. (G): Levels of CD274/PD-L1 as detected by RNAseq in normal and tumor epithelium, respectively. (H): Levels of CD274/PD-L1 as detected by qRT-PCR in normal and tumor epithelium, respectively. P-values were calculated using student's t test with significance cutoff set at P = 0.05. (I): Levels of CD152/CTLA-4 as detected by RNAseq in normal and tumor epithelium, respectively. COSCC, canine oral squamous cell carcinomas; HNSCC, head and neck squamous cell carcinomas.