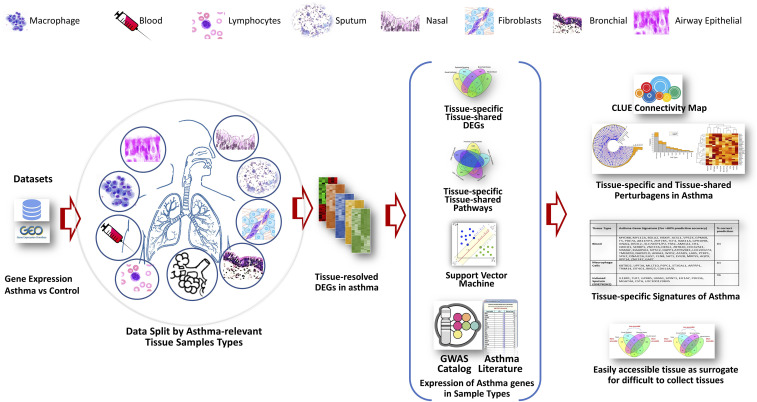
Figure 7.
Diagramatic summary of the current study starting from the discovery of differentially regulated genes to drug repositioning in asthma using multi-tissue transcriptomic analysis. Using asthma transcriptome data identified from multiple sample types such as blood, lung, isolated fibroblasts, nasal epithelium, lymphocytes, and airway macrophages we identified molecular signatures of asthma. The signatures were further connected with GWAS-identified asthma genes. The tissue-resolved molecular signatures were further evaluated for their utility in drug repurposing (i.e. identifying perturbagens via connectivity map analysis to reverse asthma signature).