Figure 10.
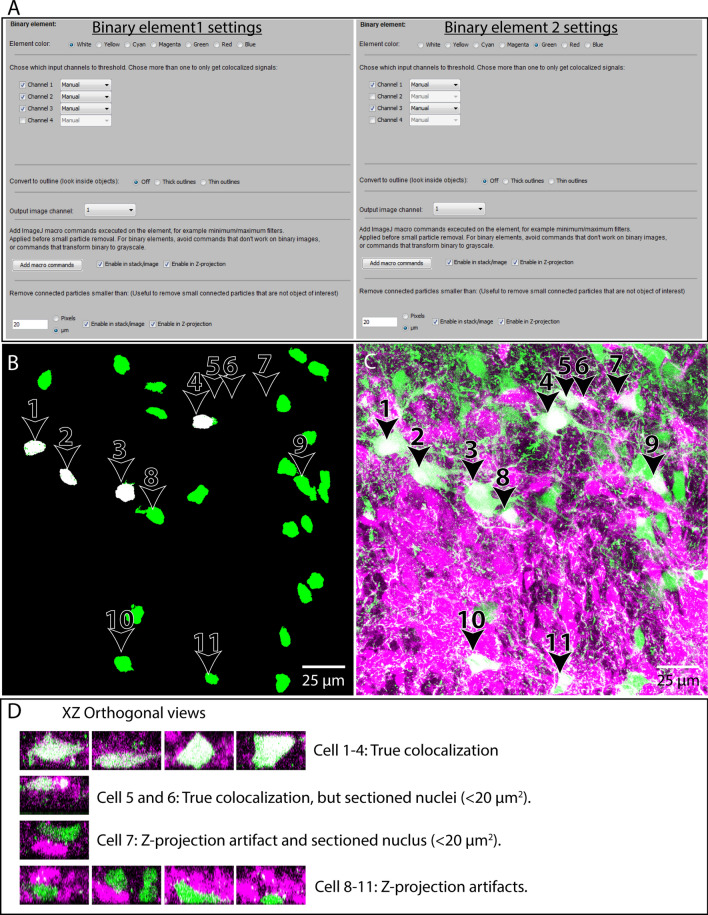
Example of usage 1. Visualizing Colocalization between two neuronal populations using the Colocalization Image Creator. Overview of the element settings (A) used with the Colocalization Image Creator plugin, and the Z-projection output (B). Three channels of the input image (see Fig. 9) are used: channel 1 showing nuclear staining (Hoechst 33258), channel 2 showing neurons labeled with tdTomato expression, and channel 3 showing retrogradely labeled neurons. (B) Z-projection output using the element settings in (A). White objects represent colocalization of nuclear, retrograde, and tdTomato labeling. Green objects represent colocalization of nuclear and retrograde labeling only. Arrowheads and numbers in (B) indicate the neurons shown in (C,D). (C) Standard maximum intensity Z-projection of the grayscale data of the two neuronal populations, using the built-in ImageJ tool (Stacks- > Zproject- > Maximum). Retrogradely labeled neurons are green, tdTomato-expressing neurons are magenta. White pixels indicate overlap, either due to true colocalization or Z-projection artifacts. (D) XZ orthogonal views of the neurons indicated in (B,C), which are colored as in (C). The different rows show examples of true colocalization, Z-projection artifacts, and neurons whose nuclei have been transected during tissue sectioning. Note how only neurons 1–4 have true colocalization of signal in all three input channels, and are completely included in the section, which is clearly shown in (B). Note also that the images in (C, D) are not outputs of the Colocalization Image Creator plugin, but only presented to evaluate the efficiency of the Z-projection artifact removal.