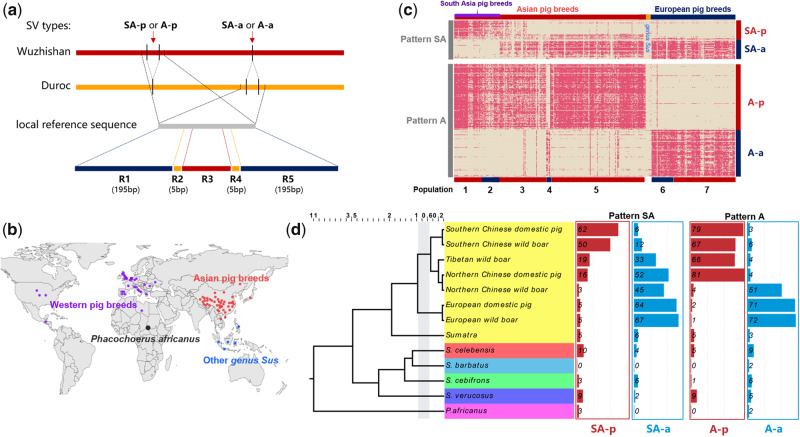
Fig. 2.
Verification of 327 specific pattern-related SVs using large-scale NGS data. (a) The local assembly methods to validate the pattern-related SVs for each pig. Five regions were defined for each SVs: R1 (left 195 bp from R2 region), R2 (left 5 bp from SV breakpoint), R3 (SV boundaries), R4 (right 5 bp from SV breakpoint), and R5 (right 195 bp from R4 region). (b) The 316 individuals from 65 different pig breeds distributed throughout Eurasia. (c) A heatmap for pattern-related SVs among 316 individuals from four major areas. The red highlight represents the SV region of a specific individual that was successfully covered by its sequencing reads. For example, A-a type with pattern A which can be covered by the sequencing reads of a European breed is marked in red; however, the sequence of A-a type with pattern A which cannot be covered by the sequencing reads of an Asian breed is not marked. Populations 1–7 represent Southern Chinese wild boar, Southern Chinese domestic pig, Tibetan wild boar, Northern Chinese wild boar, Northern Chinese domestic pig, European domestic pig, and European wild boar, respectively. (d) Phylogenetic tree of different species within the genus Sus, where bar plots indicate the percentage of the pattern-related SVs within different pig species/breeds.