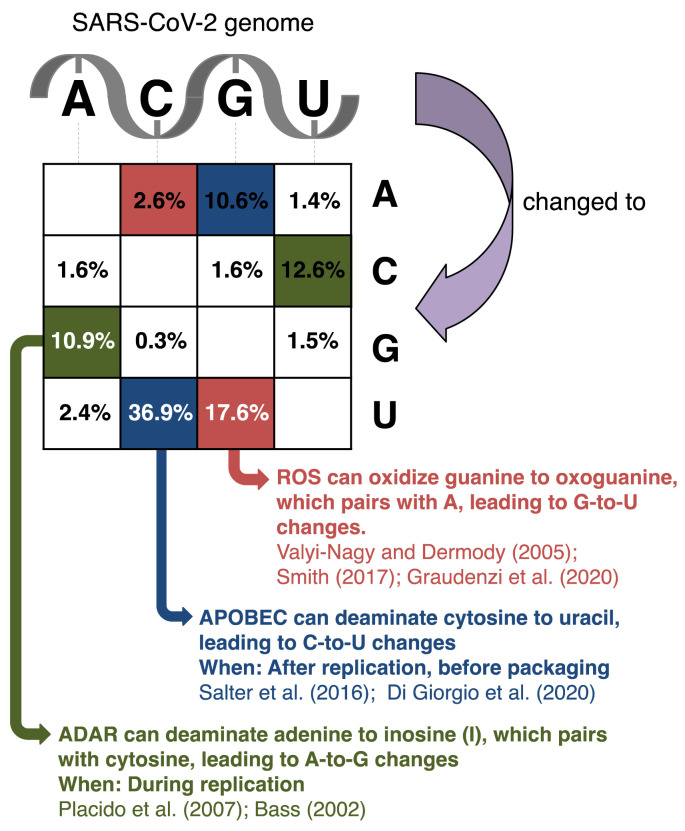
Fig. 1.
Matrix showing the distribution of genomic changes in SARS-CoV-2 sequences deposited at GISAID (https://www.gisaid.org/; [71]) as of October 2nd, 2020. Changes are accumulated across 79,887 samples and mapped onto the reference SARS-CoV-2 genome sequence, and the percentages of changes were recorded as previously described [21]. Changes at individual sites may therefore represent multiple independent events, and the most prominent changes are most likely underestimated [21]. The three types of changes resulting from the activity of ROS, APOBEC, and ADAR (G-to-U, C-to-U, and A-to-G, see main text) are highlighted in red, blue, and green, respectively. The types of changes that would result of the same host factors acting on the complement strand on double-stranded RNA are similarly colored but in a checkered pattern.