Figure 4. Modeling HES1-mediated organ segregation error in HBPOs.
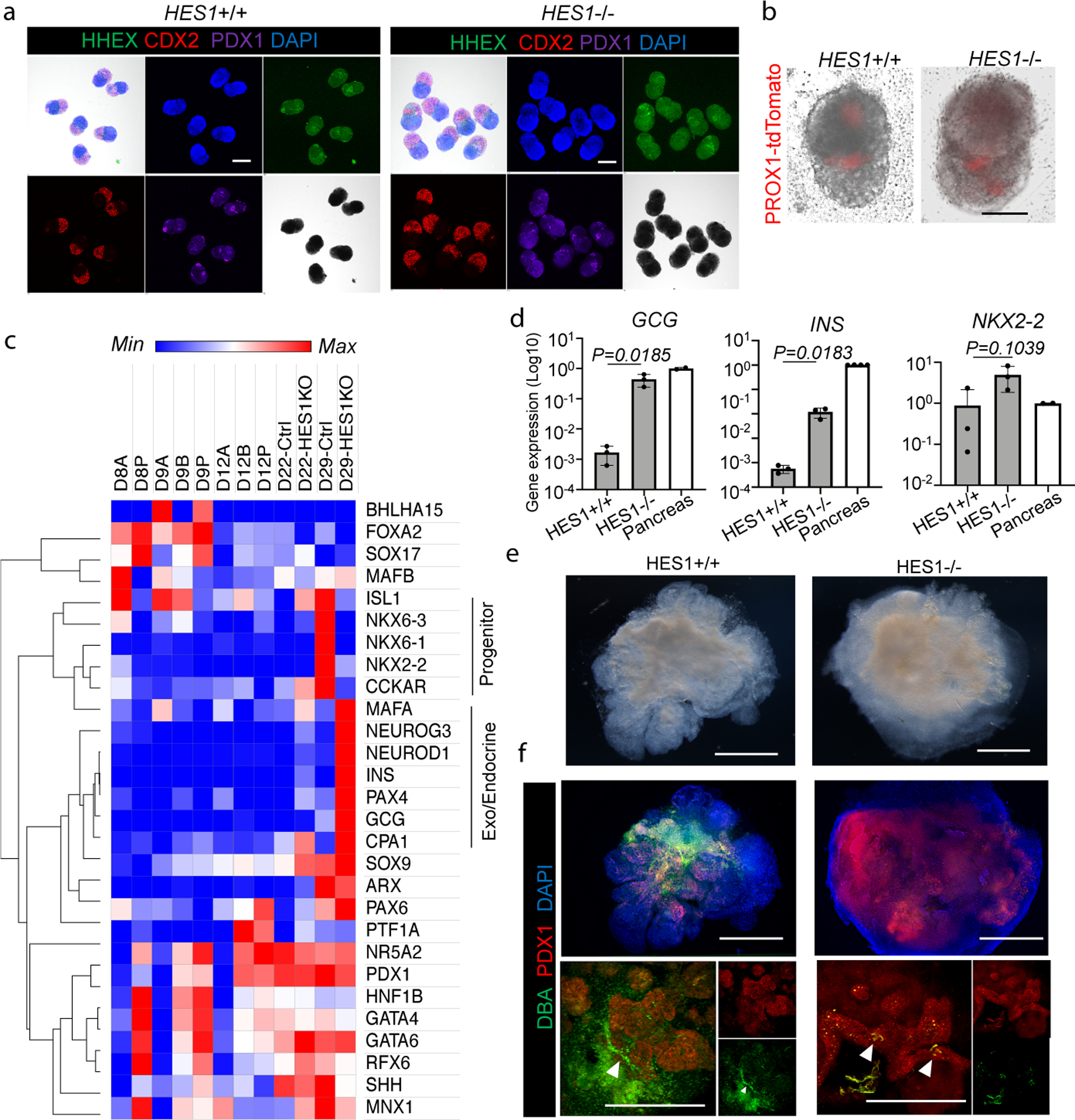
a. Confirmation of boundary domain formation from HES1 −/− iPSC by wholemount immunostaining of SOX2, CDX2, PDX1 and HHEX. Independent experiments were repeated twice with similar results.
b. PROX1-tdTomato expression in HES1 −/− organoids at D11. Representative of n = 6 independent organoids showing similar results.
c. RNAseq of pancreas associated markers at day22 of HES1 −/− HBPOs.
d. Log fold gene expression of GCG, INS, and NKX2–2 in HES1 −/− organoid and human adult pancreas. Data are mean ± s.d. n = 3 independent experiments for organoids. Unpaired, two-tailed t-test for HES1 +/+ and −/− organoids.
e. Macroscopic observation of boundary domain of HES1−/− organoid. Representative of n = 3 independent organoids showing similar results.
f. Wholemount staining for PDX1 and DBA in HES1 KO organoids. Representative of n = 3 independent organoids showing similar results.
Scale bars, 200 µm (a), 100 µm (b), 500 µm (e, f).
