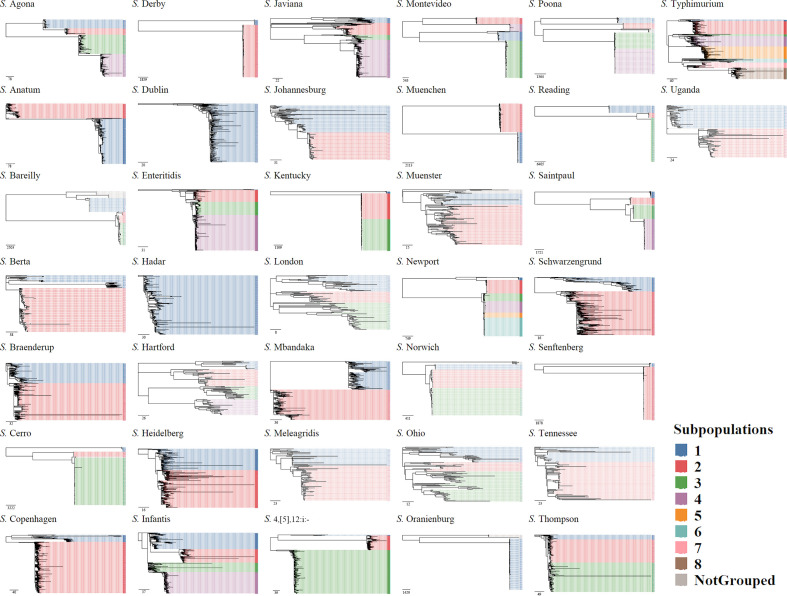
Fig. 1.
Approximate maximum-likelihood phylogenetic trees were reconstructed with FastTree using SNPs found in the core genomes of the 37 Salmonella serotypes. For each serotype, a core genome alignment was created including two S. Paratyphi type A outgroup strains (SRR3033248, SRR3277289; not included in the figure). Bootstrap replicates (n=5000) were used for branch support. Tree tips were coloured according to the identified genetic subpopulations. The scale bar indicates SNP difference.