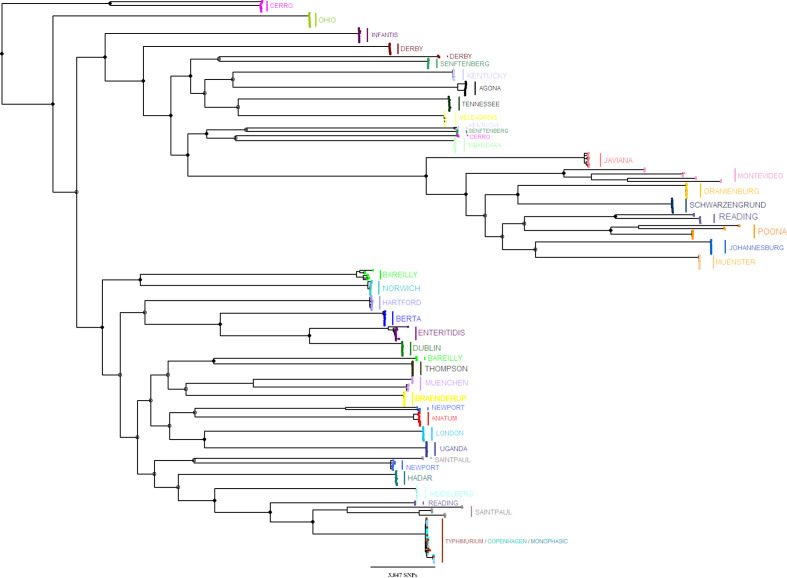
Fig. 3.
An ML phylogenetic tree was reconstructed with RAxML using SNPs found in the core genome of representative sequences from all 37 serotypes (n=370). Ten sequences were selected from each serotype phylogeny to represent the diversity of the genetic subpopulations. The tree was rooted using S. Paratyphi type A outgroup strains (SRR3033248, SRR3277289; not included in the figure). Bootstrap replicates (n=5000) were used for branch support. Full and empty circles indicate ≥70 and <70 % bootstrap support for major branches, respectively. Tree tips were coloured according to the serotype (Fig. S3 includes the same tree, annotated by the serotype and the genetic subpopulations).