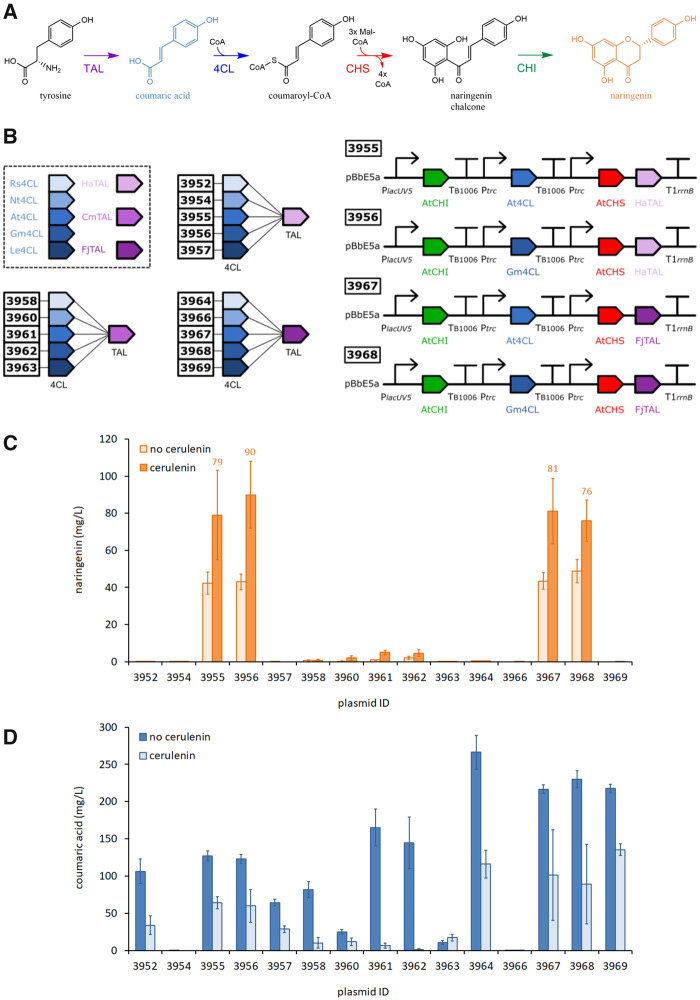
Fig 1:
Enzyme selection and naringenin pathway construction. (A) Pathway highlighting key enzyme steps from tyrosine to naringenin. Enzyme abbreviations: TAL, tyrosine ammonia-lyase; 4CL, 4-coumarate-CoA-ligase; CHS, chalcone synthase; CHI, chalcone isomerase. (B) Construction of 15 Naringenin pathways with their corresponding plasmid IDs. Enzyme abbreviations: Rs4CL, 4CL from R. sphaeroides; Nt4CL, N. tabacum; At4CL, A. thaliana; Gm4CL, G. max; Le4CL, L. erythrorhizon; HaTAL, TAL from H. aurantiacus; CmTAL, C. metallidurans; FjTAL, F. johnsoniae. The four best-performing plasmid constructs are shown. (C) Screening a library of 15 pathways for the production of naringenin in the presence or absence of cerulenin. (D) Coumaric acid titers quantified for the same constructs. Data represent the mean and standard deviation from four replicate cultures (wildtype MG1655 cells grown at 30°C for 24 h in LB media with 0.4% glycerol and 3 mM tyrosine).