Figure 1. Specific interactions with acetylated targets and Sox DNA motifs are needed for Brd4 and Sox2 clustering, respectively, while IDRs are dispensable.
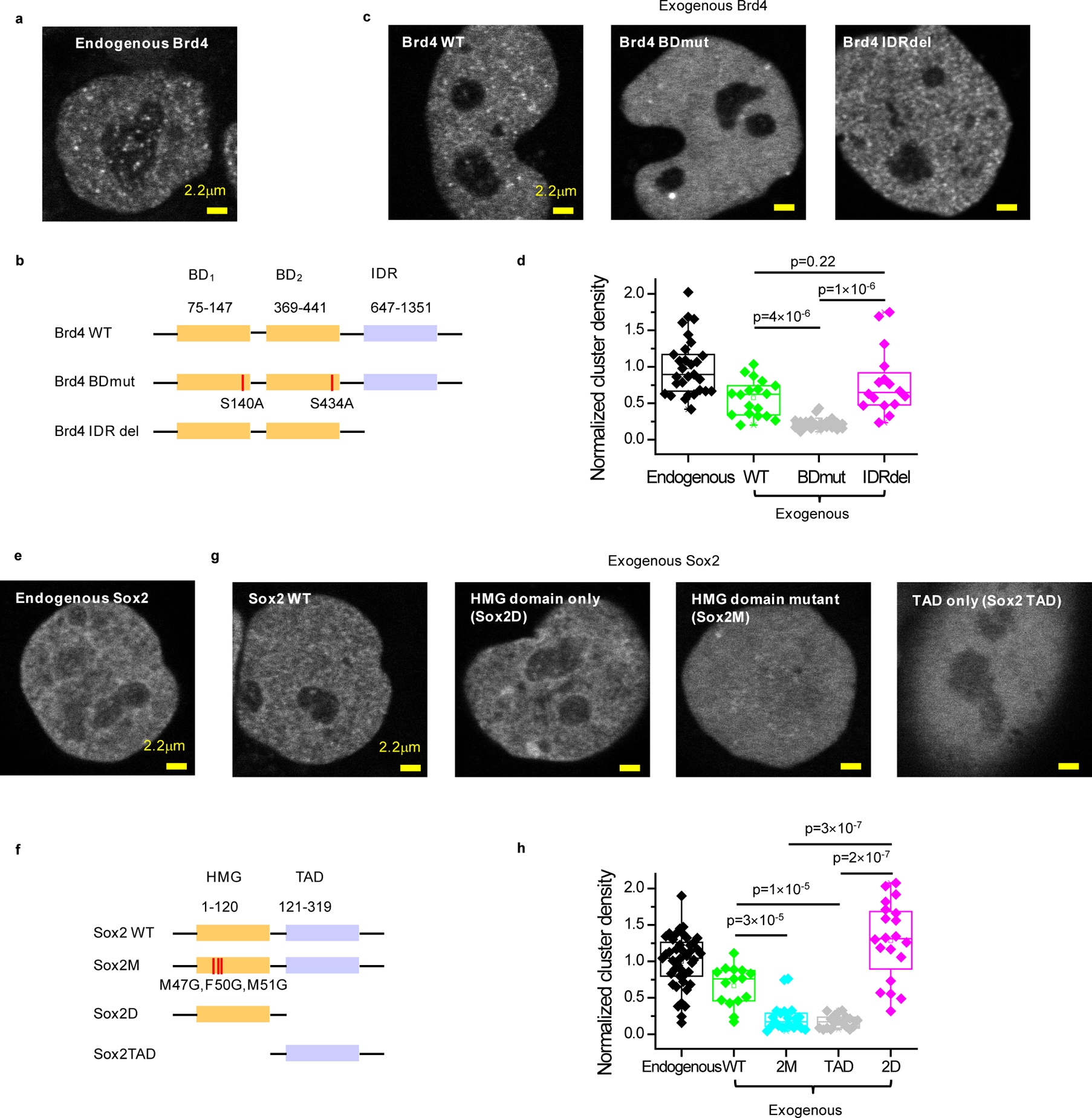
(a) Representative clustering pattern of endogenous Brd4 in live mESC nuclei. Endogenous Brd4 is visualized by bi-allelic knock-in of an N-terminal SNAP-tag and labeling with SiR-BG dye. (b) Domain organization of Brd4 and schematic of double bromodomain and IDR deletion mutants. Two serine residues (S140, S434) are mutated to alanine to generate the double bromodomain mutant, Brd4 BDmut. The entire IDR domain (647–1351) was deleted to generate the IDR deletion mutant, Brd4 IDRdel. (c) Representative images of nuclear distribution of exogenous SNAP-tag WT, BDmut and IDRdel Brd4 labeled with SiR-BG dye in live mESCs. (d) Quantification of Brd4 clustering in mESCs. For ectopically expressed WT, BDmut and IDRdel Brd4, cells with close-to-endogenous expression level are selected for analysis (Extended Data Figure 1a). Each data point corresponds to the nuclear cluster density of a single cell, normalized to the mean nuclear cluster density of endogenous Brd4. Data points are from 2 independent experiments with total n= 29, 18, 21, and 16 cells for endogenous, WT, Bdmut, and IDRdel, respectively. p-values are calculated based on a Wilcoxon rank-sum test. (e) Representative clustering pattern of endogenous Sox2 in live mESC nuclei. Endogenous Sox2 is labeled with an N-terminal SNAP-tag bi-allelic knock-in and visualized with SiR-BG dye. (f) Domain organization of Sox2 and schematic of HMG point and deletion mutants as well as TAD deletion mutant. Three residues in the HMG domain responsible for DNA binding (M47, F50 and M51) are mutated to glycine to generate Sox2M mutant. The entire TAD or HMG domain is deleted to generate Sox2D or Sox2 TAD mutants, respectively. (g) Representative images of nuclear distribution of exogenous SNAP-tag WT, 2M, TAD, and 2D Sox2 labeled with SiR-BG dye in live mESCs. (h) Quantification of Sox2 clustering in mESCs. For ectopically expressed WT, 2M, TAD, and 2D Sox2, cells with close-to-endogenous expression level are selected for analysis (Extended Data Figure 1b). Each data point corresponds to the nuclear cluster density of a single cell, normalized based on the average nuclear density for endogenous Sox2. Data points are from 2 independent experiments with total n= 43, 15, 21, 18, and 20 cells for endogenous, WT, 2M, TAD, and 2D, respectively. p-values are calculated based on a Wilcoxon rank-sum test. In panels (d, h), nuclear cluster density quantification is based on a single-cluster detection significance of p<0.001 (Extended Data Fig. 1c–d). Box-plots in (d, h): boxes indicate inter-quartile range (IQR: 25%75% intervals) and the median line, whiskers indicate 1.5× the IQR; ‘×’ symbols indicate 1% and 99% percentiles; square symbols indicate the mean. Data for graphs in (d, h) are available as source data online.
