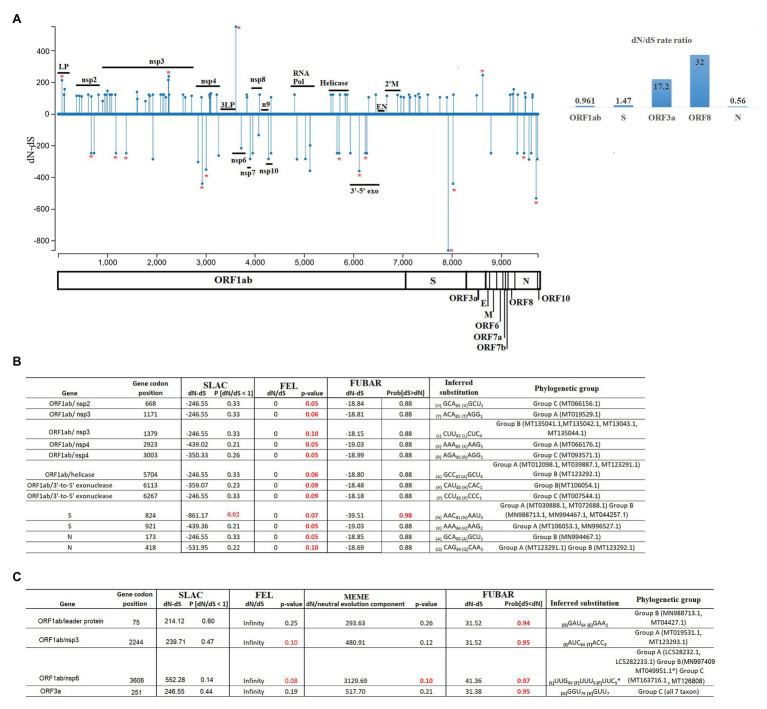
Figure 4.
Diversifying and purifying selection on SARS-CoV-2. (A) General overview obtained by SLAC analysis, showing the evolutionary rate (dN-dS or dN/dS) along the genome and at individual genes of SARS-CoV-2. Statistically significant codons were inferred by multiple evolutionary tests used in this study. Red asterisks represent codons with significant evidence for selection. Codons evolving at (B) purifying (negative) or (C) diversifying (positive) selection are shown numbers in red represent evolutionary tests with significant values according to the analysis: SLAC, FEL, MEME (p = 0.1), and FUBAR (posterior probability = 0.9). The criteria for considering a site positively or negatively selected was based on their identification by at least one of the tests. The phylogenetic group column (assigned according with Figure 2A) shows also the isolates carrying the substitutions. LP, leader protein; 3LP, 3C-like proteinase; n9, nsp9; 3'-5' exo, 3' to 5' exonuclease; EN, endoRNAse; and 2'M, 2'-O-ribose methyltransferase.