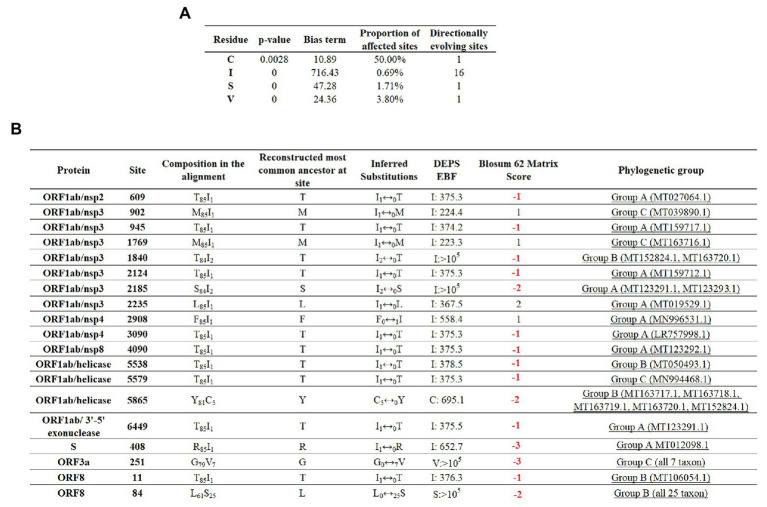
Figure 5.
Directional selection analysis on SARS-CoV-2. (A) An amino acid alignment was evaluated by DEPS and four different residues producing 19 directionally evolving sites in the proteome of SARS-CoV-2 are reported. Values of p show the statistical significance of each residue considering a model test of selection vs. not selection. Bias term: alignment-wide relative rate of substitution toward target residue. Proportion of affected sites: percentage of sites evolving under a directional model vs. a standard model with no directionality. Directionally evolving sites: number of sites that show evidence of directional selection for focal residue. (B) Description of 19 directionally evolving sites. Sites were detected by Empirical Bayesian Factor (EBF) considering a cut-off of 100 or more. Numbers in red represent replacements between amino acids with different properties. The phylogenetic group column (assigned according with Figure 2A) shows also the isolates carrying the substitutions.