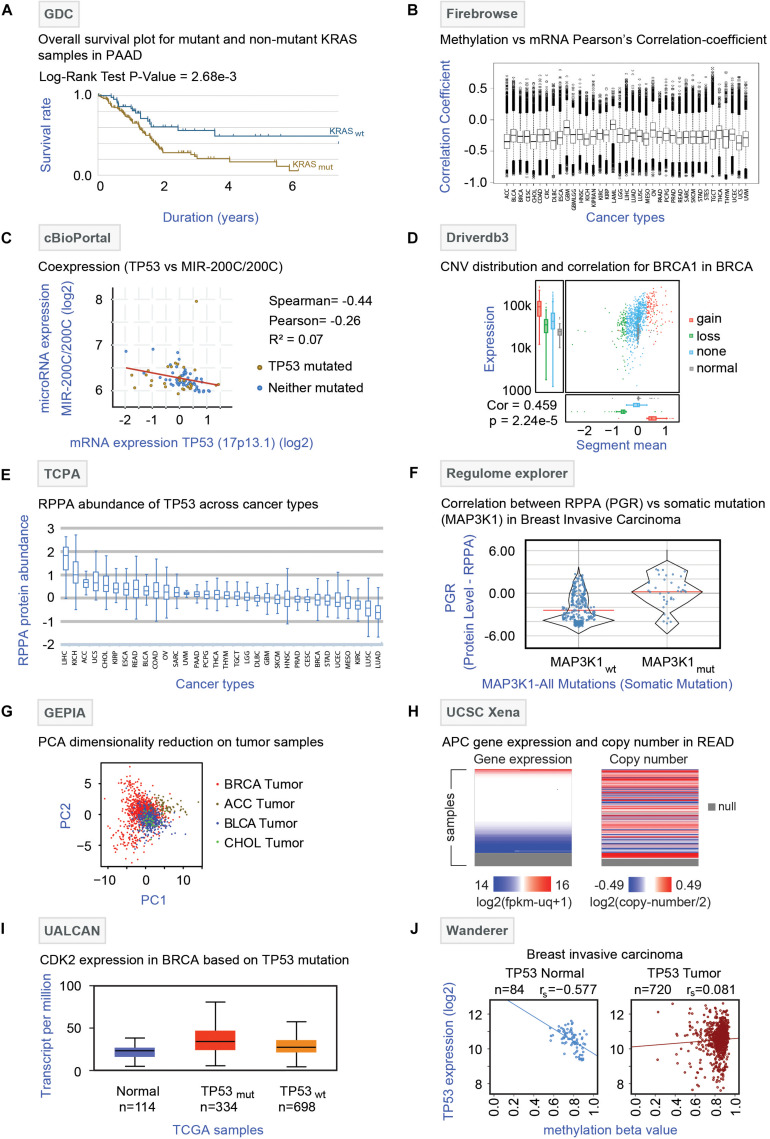
FIGURE 4.
Analysis of TCGA multi-omics data by online tools. (A) GDC analysis: Survival analysis (Kaplan Meier curve) of pancreatic ductal adenocarcinoma (PAAD) patients with (KRASmut, brown curve) or without (KRASwt, blue curve) KRAS gene mutations is shown with log-rank P-value (2.68 × 10–3). (B) Firebrowse analysis: The differential correlation pattern between methylation (beta values) and mRNA expression (TPM) was obtained from Firebrowse and shown as a box plot across 37 different TCGA cancer types. (C) cBioPortal: Correlation plot depicting the correlation between the TP53 gene and microRNA-200C expression is shown. The red line represents the regression slope. Spearman, Pearson, and Kendall rank correlation coefficients are given. (D) Driverdb3: Distribution of copy number variation (CNV) of the BRCA1 gene in breast cancer (BRCA) samples is shown. The colors represent different forms of the BRCA1 gene with or without CNV where red, green, blue, and gray indicate gain, loss, no-CNV, and normal, respectively. The box plots (left side) indicate the mRNA expression pattern of the different forms of BRCA1. The correlation between CNV and mRNA expression is given for the BRCA1 gene. (E) TCPA: Differential abundance profiles of TP53 protein across 32 TCGA cancer types are shown as box plots. The Y-axis indicates the protein abundance as measured by the RPPA technique. The error bars represent the variation among the samples for a particular cancer type. (F) Regulome Explorer: The comparison of the protein abundance of progesterone receptor (PGR) between breast cancer (BRCA) patients with (MAP3K1mut) or without (MAP3K1wt) MAP3K1 mutation is shown. The Y-axis represents protein abundance as identified by the RPPA technique. (G) GEPIA analysis: Dimensionality reduction through principal component analysis is shown. Four different tumor samples – BRCA, breast carcinoma; ACC, adrenocortical carcinoma; BLCA, bladder carcinoma; and CHOL, cholangiocarcinomas were plotted in two-dimensional planes of component 1 (PC1) and 2 (PC2) based on the mRNA expressions of 20 genes (STARD3, CASC3, CLNS1A, MED1, LSM1, RPL19, BRF2, RPS6KB1, FADD, PSMD3, UBE2Z, PPFIA1, GRB7, RSF1, HEATR6, SNF8, ERBB2, ASH2L, WHSC1L1, and PHB) for which the highest correlation between mRNA expression and methylation was found for breast cancer. The color represents each of the cancer type (Red: BRCA, brown: ACC, Blue: BLCA, and green: CHOL). (H) UCSC Xena: Gene expression and copy number alteration of APC gene in tumor samples of rectum adenocarcinoma (READ) patients are shown. The null section indicates a lack of data for those samples. (I) UALCAN: mRNA expression of the CDK2 gene were plotted for normal and the breast cancer (TP53 mutant and TP53 non-mutant) samples. (J) Wanderer: TP53 expression and methylation beta values were plotted for normal (N = 84) and breast cancer (N = 720) tissues. The correlation coefficient (rs) for each plot is indicated.