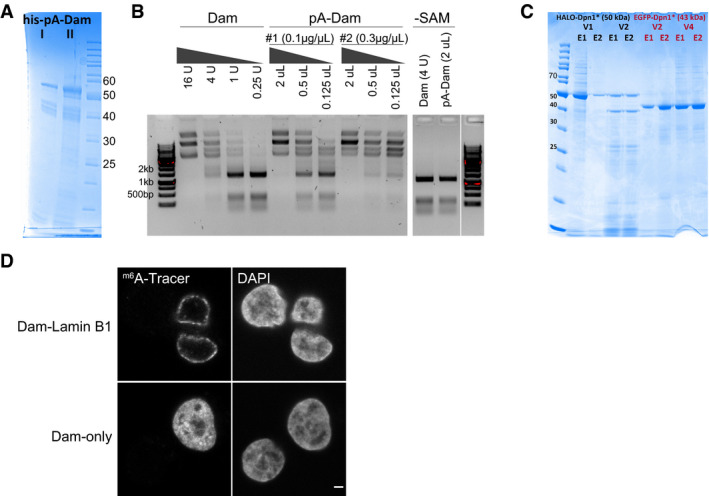
Figure EV1. Production and validation of the pA‐DamID proteins.
-
ASDS–PAGE gel of two batches of bacterial purified pA‐Dam protein (expected molecular weight: 52 kDa) with concentrations of ˜0.1 and ˜0.3 μg/μl, respectively.
-
BAgarose gel analysis of Mbo I protection assay. Unmethylated plasmid is m6A methylated by a range of Dam and pA‐Dam concentrations. The DNA is subsequently digested by Mbo I, which can cut GATC but not Gm6ATC sequences. The pA‐Dam batches have Dam activities estimated to be ˜8 and ˜32 units (see Materials and Methods for definition) per ˜0.1 μg and ˜0.3 μg pA‐Dam protein, respectively. No Mbo I protection is observed without the methyl donor SAM.
-
CSDS–PAGE gel of m6A‐Tracer protein purified from insect cells, with the truncated DpnI fused to EGFP and HALO tags. Two elutions (labeled E1/E2) obtained with two independent baculovirus pools (labeled V1/V2 and V2/V4) produced protein of the expected size (50 and 43 kDa for EGFP and HALO‐tagged protein, respectively) and were pooled.
-
DConfocal sections of m6A‐Tracer signal in HAP‐1 cells transduced with lentivirus expressing Dam‐Lamin B1 (top panel) and Dam‐only (bottom panel). Negative cells are presumably non‐transduced cells. The scale bar corresponds to 2 μm.
