-
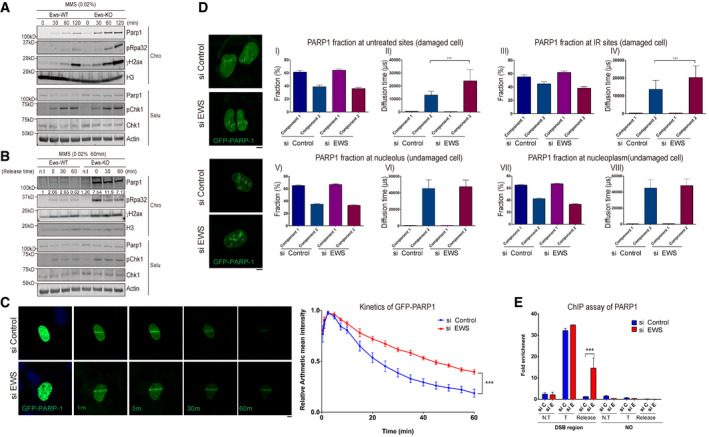
A
Western blot analysis of Parp1 and DNA damage markers in wild‐type (WT) and Ews
−/− mBA cells. Cells were treated with MMS (0.02%) in a time dependent manner. Proteins were fractionated into two groups, chromatin‐bound (Chro) proteins and soluble (Solu) proteins.
-
B
After treatment with MMS (0.02%, 1 h), the media was replaced to release the DNA damage. The proteins were fractionated and the specific protein kinetics at chromatin were analyzed by Western blot.
-
C
GFP‐PARP1 U2OS cells were transfected with siControl and siEWS. Local DNA damage was induced by micro‐irradiation using a 405 nm laser. Data represented as mean ± SEMs and more than six cells were analyzed from six independent experiments. The statistical significance determined by one‐way ANOVA, ***P < 0.001. Scale bar indicates 5 μm.
-
D
Fluorescence correlation spectroscopy (FCS) was measured after 15 min later following micro‐irradiation (Upper) or under normal condition in GFP‐PARP1 U2OS cells with either siControl or siEWS. Data represented as mean ± SEMs, more than 50 cells were analyzed, and significance determined by one‐way ANOVA, ***P < 0.001. Scale bar indicate 5 μm.
-
E
AsiSI endonuclease‐integrated U2OS cells were transfected with siControl and siEWS. Cells were incubated with doxycycline for 4 h to induce DSBs. With or without changing the Dox‐added media to fresh media for 2 h (for release samples), the amount of chromatin‐associated PARP1 was measured using ChIP assay. N.T: Non‐treat, T: AsiSI treat, Release: Damage released samples. Data represented as mean ± SEMs, and technical repeats (n = 3), significance determined by two‐way ANOVA, ***P < 0.001.