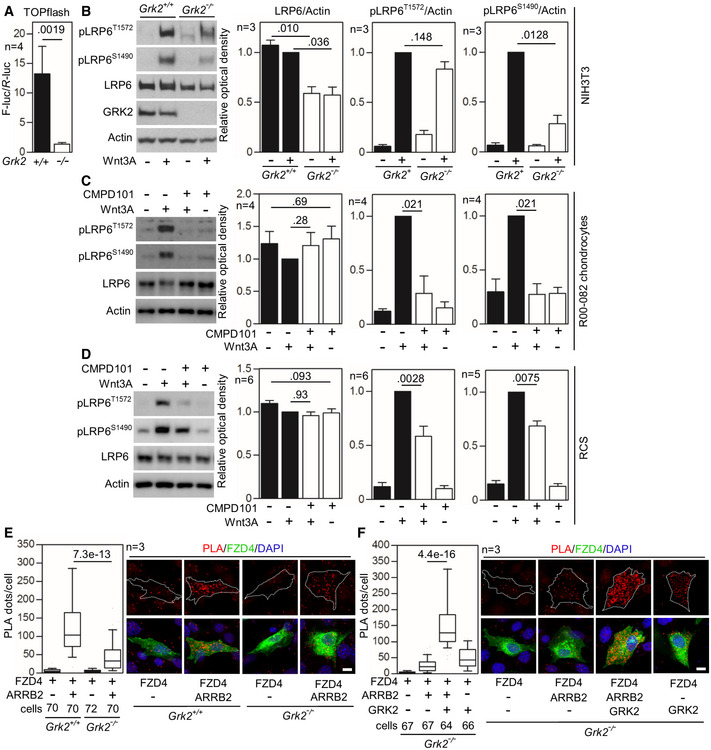
Figure 7. Inhibition of GRK2 activity inhibits canonical Wnt signaling.
-
AControl (+/+) and Grk2 −/− NIH3T3 cells were transfected with the TOPflash Firefly (F) luciferase vector together with a control Renilla (R) luciferase vector, treated with Wnt3A (100 ng/ml) for 24 h and subjected to dual‐luciferase assay. Note the impaired response to Wnt3A in Grk2 −/− cells. Mean ± SEM. Mann–Whitney U test; number of biological experiments is indicated.
-
BControl (+/+) and Grk2 −/− NIH3T3 cells were treated with 40 ng/ml Wnt3A for 1 h. Note the lower LRP6 expression and inhibited Wnt3A‐induced LRP6 phosphorylation in the Grk2 −/− NIH3T3 cells. Mean ± SEM. Welch's t‐test; number of biological experiments is indicated.
-
C, DLoss of LRP6 phosphorylation in chondrocytes treated with the GRK2 inhibitor CMPD101. Control human R00‐082 chondrocytes (C) and rat chondrosarcoma (RCS) cells (D) were treated with 20 μM CMPD101 overnight and then treated with 100 ng/ml Wnt3A for 1 h. Note the less LRP6 phosphorylation (pS1490‐ and pT1572‐LRP6) in cells treated with CMPD101. Mean ± SEM. Mann–Whitney U test; number of biological experiments is indicated.
-
E, FLoss of interaction between Frizzled 4 (FZD4) and ß‐Arrestin 2 (ARRB2) in Grk2 −/− NIH3T3 cells. (E) Proximity ligation assay (PLA) between expressed FZD4‐GFP and ARRB2‐Flag showed significantly reduced proximity events between the two proteins in Grk2 −/− cells. (F) GRK2 add‐back rescued the interaction. White dashed lines outline the transfected cells. Central band, median. Box, 1st–3rd quartile. Whiskers, 10–90% percentile. Mann–Whitney U test; number of biological experiments and the total numbers of analyzed cells are indicated. Scale bars, 10 μm.
Source data are available online for this figure.
