Fig. 2. Biotin synthesis gene clusters containing putative bioZ genes and alignment of BioZ proteins with E. coli FabH.
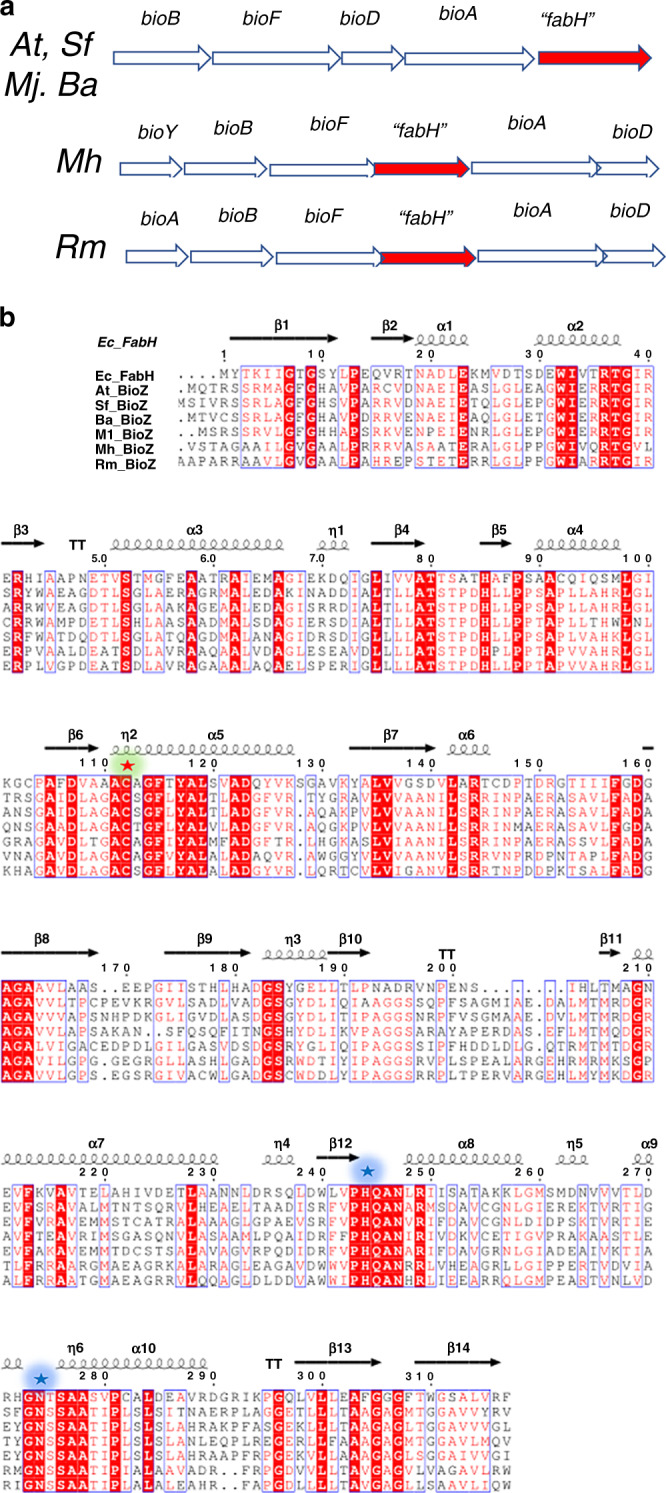
a The annotated biotin gene clusters of four species of α-proteobacteria are shown where “fabH” is bioZ. At is Agrobacterium tumefaciens, a plant pathogen; Sf is Sinorhizobium fredii, a plant symbiont; Mj is Mesorhizobium japonicum (Mj), formerly Mesorhizobium loti (Ml), another plant symbiont and Ba is Brucella abortus, an animal and human pathogen. Two thermophilic bacteria, Marinithermus hydrothermalis (Mh, an atypical Bacteroidetes) and Rhodothermus marinus (Rm), also contain bioZ (“fabH”) genes. b Multiple sequence alignments of E. coli FabH and the BioZ proteins. Amino-acid residues conserved between FabH and all BioZ proteins are in red. Potential BioZ catalytic residues conserved in KAS III proteins are highlighted in green (the active site cysteine residue that carries the acyl enzyme intermediate) or blue (the essential histidine and asparagine residues critical for the decarboxylation step of the FabH reaction).
