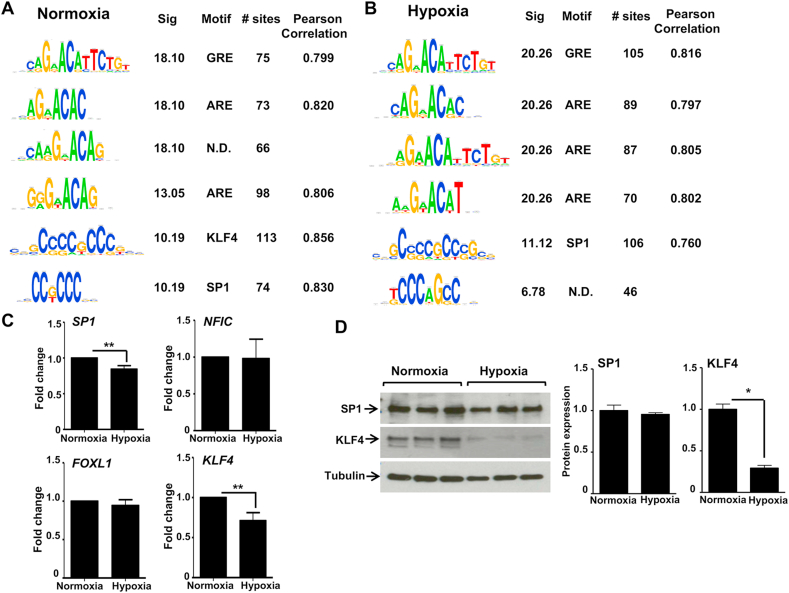
Fig. 4.
Hypoxia eliminates the enrichment of KLF4 motif at GR binding sites. (A, B) Motifs under peaks of GR binding in normoxia and hypoxia were analyzed using RSAT. The top 6 GR binding motifs identified were ranked by adjusted p value, ‘Sig’. Equally ranking motifs were ordered by decreasing percentage of GR binding regions containing the motifs. The degree of similarity between discovered motifs and matching motifs in JASPAR were measured by Pearson correlation coefficient. N.D. refers to the motifs were not discovered previously. (C) HeLa cells were cultured in either normoxia or hypoxia overnight. RNA samples were purified and the expression of SP1, NFIC, FOXL1 and KLF4 transcripts determined. (D) Protein samples were extracted from HeLa cells and KLF4 and SP1 expression measured by immunoblotting. Immunoblots show samples from three independent experiments. Immunoreactive bands for SP1 and KLF4 were quantified using Image J software, normalised to tubulin expression and then represented as a fold change over normoxia cultured cells. Graphs show mean ± S.D. of experiments repeated three times. **p < 0.01, *p < 0.05.