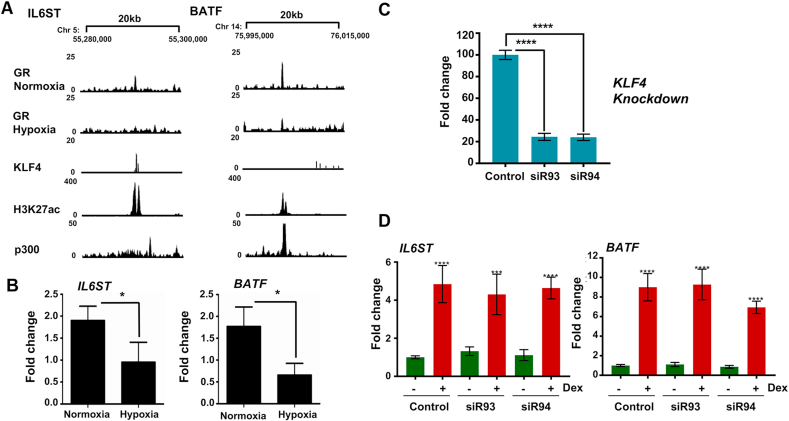
Fig. 5.
MicroRNAs regulate the impaired GR transactivation in hypoxia. (A) Cistrome for KLF4 was also aligned with GR peaks, highlighted IL6ST and BATF as genes under common regulation. Individual UCSC browser gene tracks for IL6ST and BATF are shown with GR, KLF4, H3K27ac and p300 peaks. (B) Cells were cultured in normoxia or hypoxia overnight, then treated with control or 100 nM dexamethasone for 4 h before harvest. Expression levels of either IL6ST or BATF transcript were measured by qRT-PCR. Graphs show the fold change of transcripts in response to treatment with dexamethasone compared to control. Mean ± S.D. n = 3. (C) HeLa cells were transfected with KLF4-specific siRNAs (siR93 and 94) and a negative control siRNA for 72 h. KLF4 knockdown efficiency was measured by qRT-PCR. Representative data are shown as mean ± S.E.M. n = 8. (D) Following KLF4 knockdown, cells were treated with dexamethasone (Dex) or DMSO as the vehicle. ChIP-qPCR was performed using IL6ST and BATF specific primers. CT values were normalised to Spike-In chromatin. Data are presented as fold enrichment over the Veh-treated control group. Data are shown as mean ± S.E.M. n = 4. ****p ≤ 0.0001, ***p ≤ 0.001, *p < 0.05.