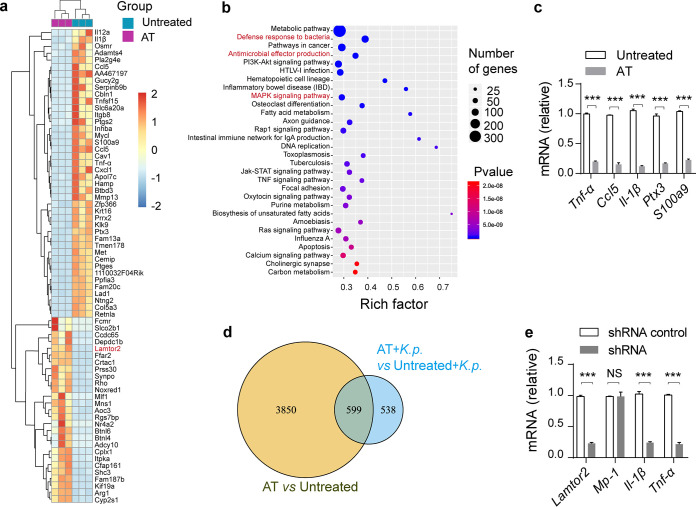
FIG 4.
Gut microbiota depletion alters alveolar macrophage transcriptome profiles. (a) Hierarchical clustering heatmap of selected significant differentially expressed genes (DEGs) in alveolar macrophages from antibiotic-treated and untreated controls. Red denotes increased expression; green denotes decreased expression. (b) Selected examples of KEGG pathway enrichment. The defense response to bacteria, antimicrobial effector production, and the MAPK/ROS signaling pathway are highlighted in red. (c) Validation of Tnf-α, Ccl5, Il-1β, Cxcl1, and S100a9 mRNA expression using RT-qPCR. (d) Venn diagram of differentially expressed genes among four groups. (e) Responsiveness of shRNA-mediated Lamtor2 knockdown RAW264.7 cells and controls in terms of cytokine and chemokine production against K. pneumoniae infection. The group size was 8 to 12 mice. Data are from three independent experiments (c, e). The P values were determined using the hypergeometric test and Benjamini-Hochberg FDR correction (b) or two-tailed Student t tests (c, e). ***, P < 0.001. NS, not significant; AT, antibiotic-treated mice.