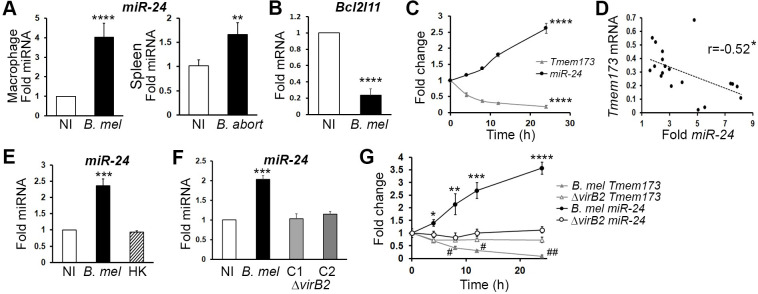
Fig 4. Brucella induces a STING-targeting microRNA miR-24.
A) Left panel: Macrophages were not infected (NI) or infected with 100 MOI Brucella melitensis (B. mel) for 24 hours before harvesting for RNA. Right: Mice were infected with 106 B. abortus 2308 (B. abort) for 24 hours prior to processing of spleen for microRNA. Micro RNA levels were determined by qPCR with normalization toRNU6 and uninfected controls (NI set = 1). In vitro results are from 17 experiments, with error bars denoting SEM. In vivo, results are representative of 2 independent experiments. N = 3 uninfected and 4 infected mice, with SD error bars. B) Macrophages were infected as in (A) and processed for mRNA. Expression was normalized to 18S rRNA. Bcl2l11 (Bim) expression is from 8 experiments. C) Time course of quantitative expression of both miR24-3p and STING (Tmem173) mRNA. P<0.001 for changes over time (N = 6). D) Correlation is from 19 experiments performed and evaluated as in (A). R2 = 0.027, p = 0.022 E) Macrophages were infected as in (A) and processed for miRNA. Comparison of live and heat killed B. melitensis (B. mel vs. HK) is from N = 9. P<0.005 for B. mel vs. NI and HK. F) Macrophages were infected with wild type B. melitensis or VirB2 deletion mutant clones C1 and C2 and analyzed as in (A). Results are from 3 experiments. P<0.005 for B. mel vs. NI and vs. ΔVirb2 clones. G) Time course comparing effects of wild type B. melitensis (filled symbols) and Δ virB2 (clone 1, open symbols) on miR-24 (black circles) and Tmem173 mRNA (gray triangles). Gene expression changes were normalized to time 0 for each Brucella genotype infection (see methods) and error bars represent standard deviations of triplicate determinations. P-values compare Brucella genotypes at each time point: #p<0.005, ##p<0.001 for Tmem173 and *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 for miR-24.