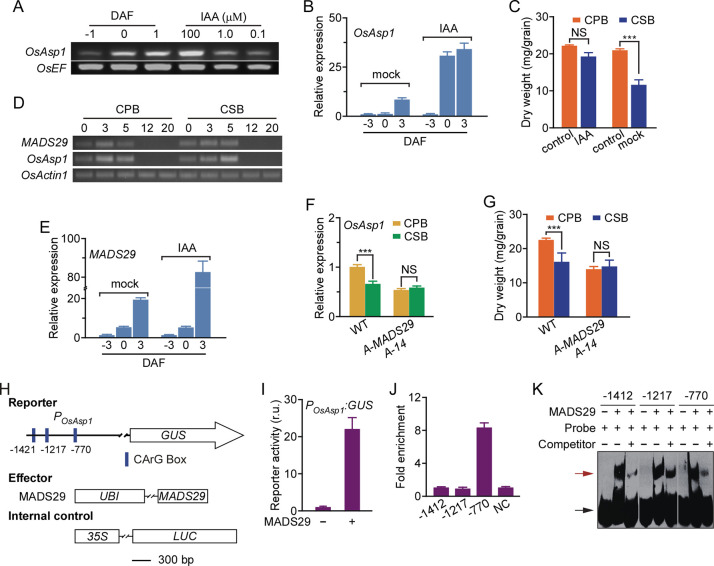
Fig 6. IAA promotes the transcription of OsAsp1 through MADS29 to maintain differential levels of OsAsp1 in CPBs and CSBs after pollination.
A, OsAsp1 levels detected by semi-quantitative RT-PCR in spikelets at –1, 0, and 1 DAF, and in detached spikelets at –1 DAF after culture in Murashige and Skoog (MS) medium plus IAA for 24 h. OsActin1 was used as a control. B, Quantification of OsAsp1 levels in CSBs of the WT treated at –3 and 0 DAF with IAA, or with methanol as a mock treatment. OsAsp1 expression was normalised to that of OsActin1, and relative expression of OsAsp1 at –3 DAF in mock-treated samples was set at 1.0. Values are means ± SD of three independent experiments. C, Mature grain dry weight of CPBs and CSBs after the IAA treatment described in B. Values are means ± SD (n = 30). NS, no significant difference; ***P < 0.001 (Student’s t-test). D, Transcript levels of MADS29 and OsAsp1 detected by RT-PCR in CPBs and CSBs of the WT at 0, 3, 5, 12, and 20 DAF. OsActin1 was used as a control. E, Transcript level of MADS29 in CSBs of the WT in response to IAA treatment. CSB spikelets on panicles were treated at –3 and 0 DAF with IAA, or with methanol as mock treatment. MADS29 expresion was normalised to that of OsActin1, and relative expression of MADS29 at –3 DAF in mock-treated samples was set at 1.0. Values are means ± SD of three independent experiments. F, Transcript levels of OsAsp1 in CPBs and CSBs of the WT and Antisense-MADS29 transgenic rice A14 (A-MADS29 A-14) at 5 DAF. OsAsp1 expression was normalised to that of OsActin1, and relative expression in CPBs of the WT was set at 1.0. Values are means ± SD of three independent experiments. NS, no significant difference; ***P < 0.001 (Student’s t-test). G, Mature grain dry weight of CPBs and CSBs of the WT and A-MADS29 A-14. Values are means ± SD (n = 30). ***P < 0.001 (Student’s t-test); NS, no significant difference. H, Constructs used for GUS assay of OsAsp1 promoter activity. The three predicted MADS29 binding sites (CArG box) were labelled in the 1500-bp promoter region of OsAsp1. LUC, firefly luciferase; GUS, beta-glucuronidase. Scale bars, 300 bp. I, Promoter activity of POsAsp1 activated by MADS29 in the protoplast of Arabidopsis leaves. POsAsp1:GUS was co-transfected with MADS29 or an empty vector as a control. 35S:LUC was used as an internal control. Relative GUS activity normalised to luciferase activity was adopted as the promoter activity of POsAsp1. Values are means ± SD of three independent experiments. J, ChIP-qPCR was used to detect the enrichment of CArG boxes by MADS29 at sites –1421, –1217, and –770 of the OsAsp1 promoter region. 35S:MADS29-GFP and POsAsp1:GUS were co-transformed into the protoplast of Arabidopsis leaves, and MADS29-GFP was purified with GFP antibody. NC, negative control. Values are means ± SD of three independent experiments. K, EMSA showing the recombinant MADS29-His directly binding the promoter region of OsAsp1 on its CArG boxes. The CArG boxes at positions –1421, –1217, and –770 were labelled with biotin as the probe, and the unlabelled DNA fragments were used as the competitor. Red and black arrows indicate the shifted band and free probe, respectively.