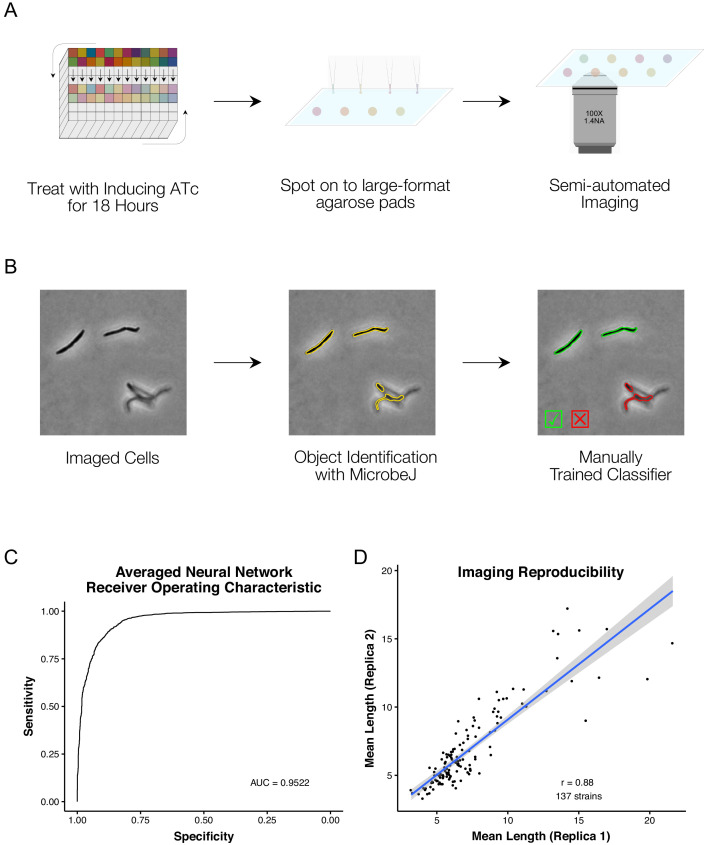
Figure 2. A high-throughput, quantitative CRISPRi-imaging pipeline for mycobacteria.
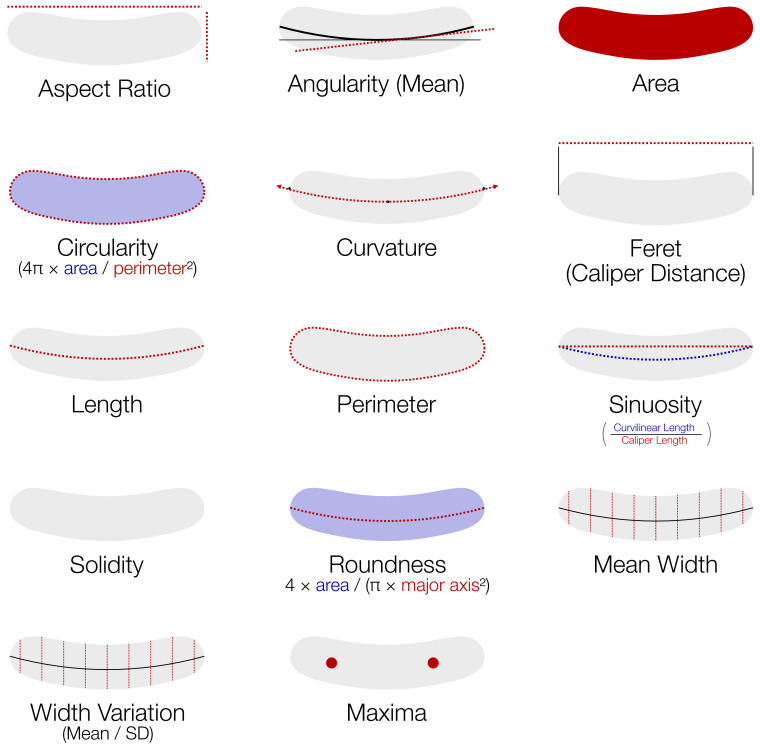
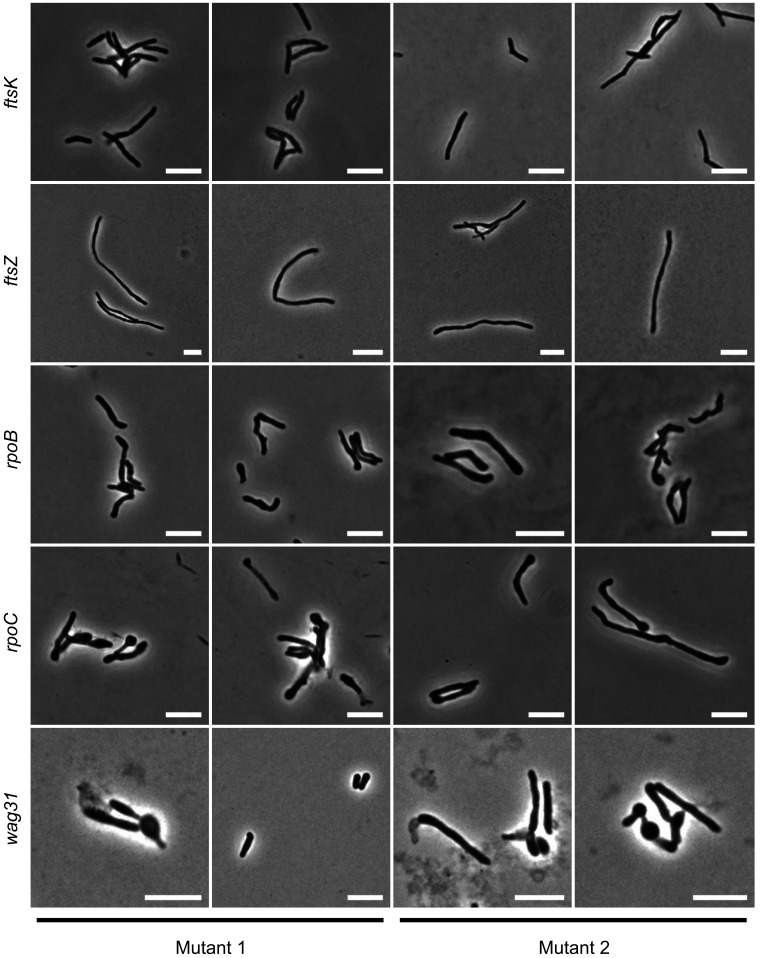
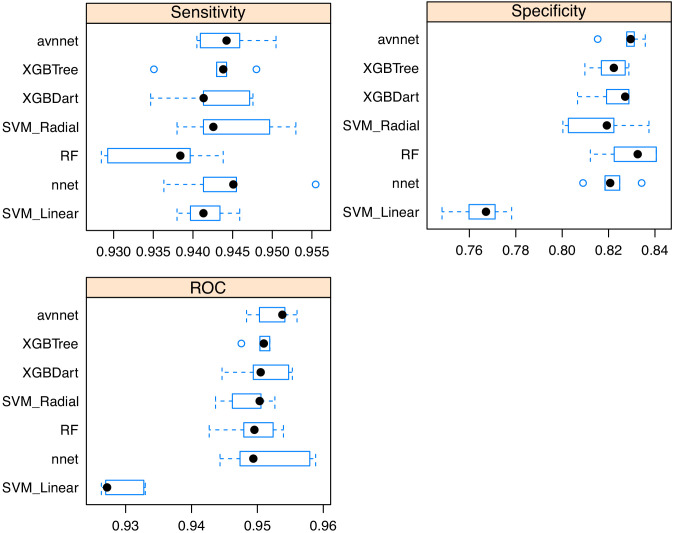
(A) Cells were exposed to ATc inducer for 18 hr before spotting onto large-format agarose pads for semi-automated imaging. (B) Image processing in MicrobeJ (Ducret et al., 2016) was combined with a manually trained Averaged Neural Network classifier to extract quantitative descriptions of bacterial morphologies and ParB protein localizations for 163559 cells across 263 gene-specific CRISPRi mutants and 27 empty vector controls. (C) Classifier performance was measured by Receiver Operating Characteristic (ROC) Area Under the Curve (AUC), returning good performance metrics. (D) Mean cell lengths were compared for 137 strains imaged as biological replicates on two separate occasions, and showed high reproducibility (r = 0.88, Pearson’s).