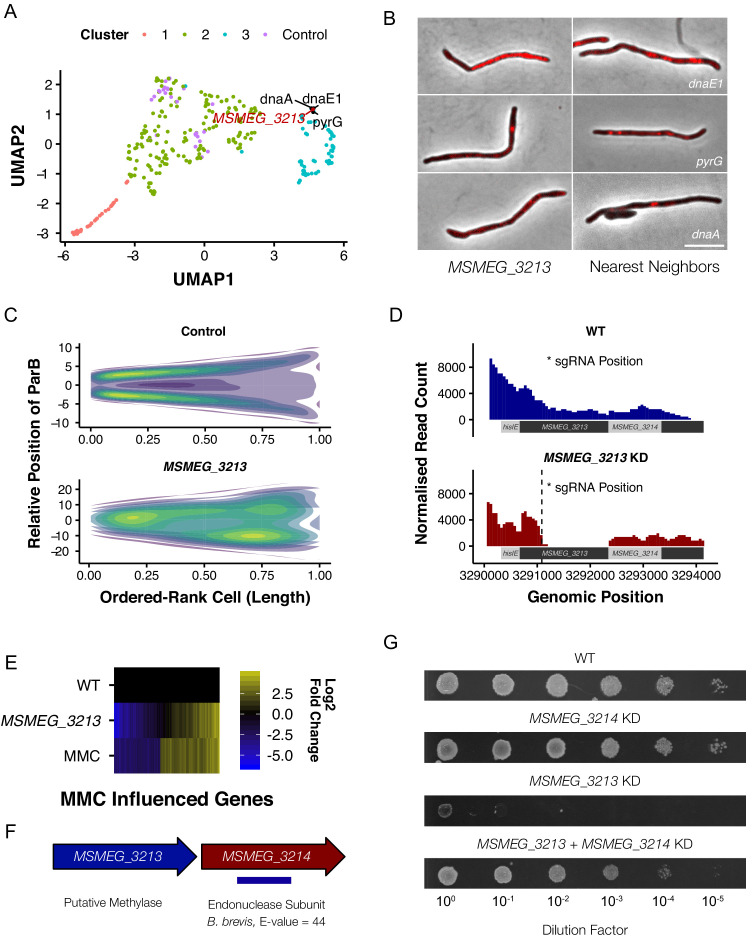
Figure 7. M. smegmatis encodes a previously undescribed restriction-modification system.
(A) MSMEG_3213, a putative DNA methylase, is associated with genes involved in DNA replication and repair in UMAP space. (B) Knockdown of MSMEG_3213 leads to cellular filamentation. The morphological nearest neighbors are dnaE1, pyrG and dnaA. (C) Consensus heatmaps of ParB-mCherry localization demonstrate that oriC positioning is disrupted in the MSMEG_3213 knockdown mutant. (D) Nanopore-based RNA-Seq confirmed that knockdown of MSMEG_3213 was specific to the targeted sgRNA position. (E) MSMEG_3213 produces a transcriptional response comparable to treatment with the DNA damaging agent, mitomycin C (MMC). (F) MSMEG_3213 is located upstream of MSMEG_3214, a gene with weak homology, according to HHpred (Hildebrand et al., 2009), to an endonuclease subunit (REBASE: 3098 BbvCI). (G) Lethality of MSMEG_3213 knockdown is suppressed by simultaneous CRISPRi-mediated knockdown of MSMEG_3214.