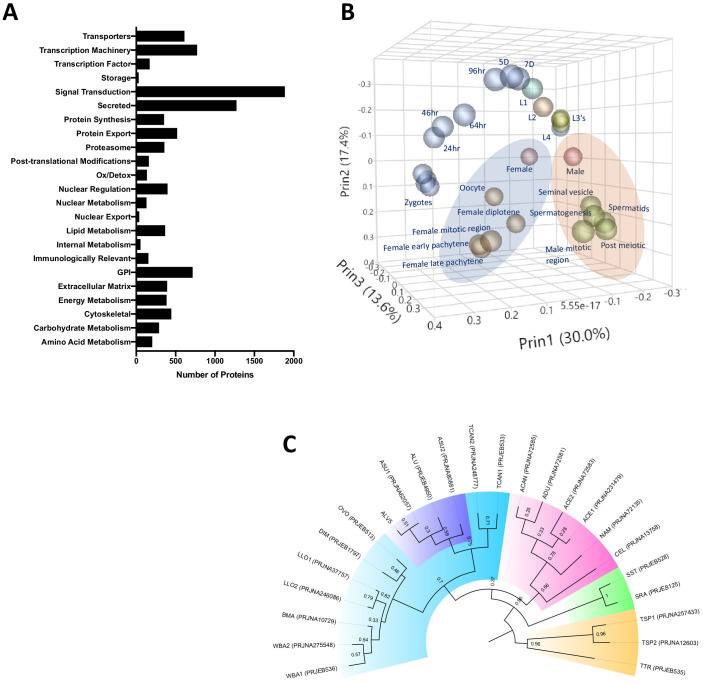
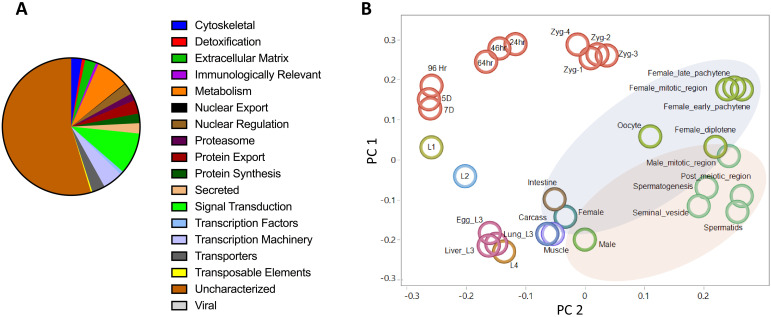
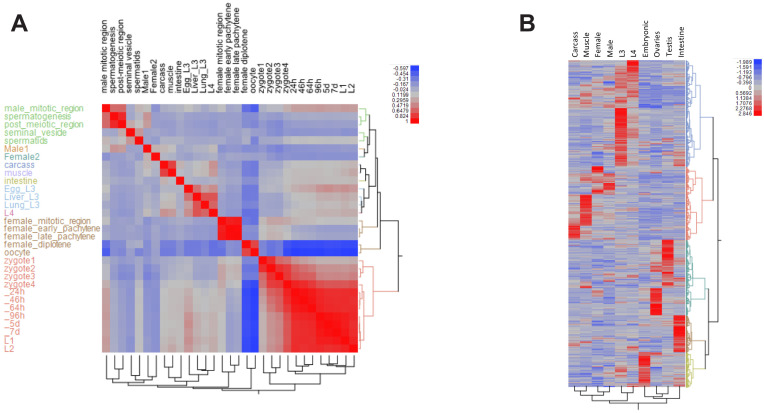
Figure 1. Ascaris proteome.
(A) Functional classification of the predicted proteome of A. lumbricoides (an improved proteome of Ascaris spp.), excluding proteins with unknown or uncharacterized function. (B) PCA plot based on multivariate analyses of RNA-seq data from various stages/tissues. Samples from tissues related to sperm (blue ellipse) and oocyte production (orange ellipse, see also Figure 1—figure supplement 2) cluster together. (C) Estimated tree based on orthology analyses between the predicted proteomes of publicly available nematodes. The Ascaris clade has been shaded in purple within Clade III (teal). Samples are labeled by BioProject Accession number, as well as by the first letter of the genus and the first two letters of the species name (ASU = Ascaris suum, ALU = Ascaris lumbricoides, WBA = Wuchereria bancrofti, BMA = Brugia malayi, LLO = Loa loa, DIM = Dirofilaria immitis, OVO = Onchocerca volvulus, TCAN = Toxocara canis, ACAN = Ancylostoma caninum, ADU = Ancylostoma duodenale, ACE = Ancylostoma ceylanicum, NAM = Necator americanus, CEL = Caenorhabditis elegans, SST = Strongyloides stercoralis, SRA = Strongyloides ratti, TSP = Trichinella spiralis, TTR = Trichuris trichiura). Multiple genomes for the same organism are suffixed with numerals.